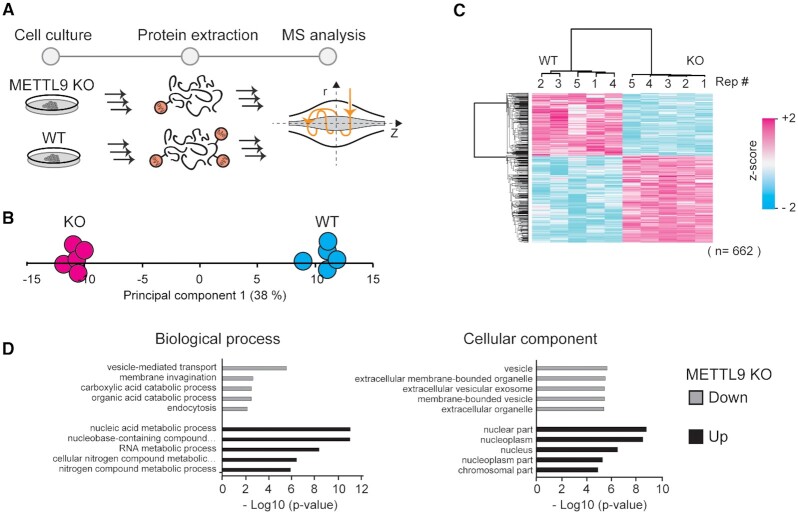
Figure 6.
Cellular effects of METTL9 knockout. (A) Workflow for proteomics characterization of METTL9 KO cells. Proteins were extracted from HAP-1 wild type (WT) and METTL9 knockout (KO) cells and processed for label free quantitative mass spectrometry. (B) Principal component analysis. Separation of experimental conditions (WT and KO) and replicates (n = 5) in the first principal component is shown. (C) Clustering analysis. Hierarchical cluster of z-scored LFQ intensities for proteins having a significant difference in abundance (Student's t-test, P-adj < 0.05; Benjamini-Hochberg FDR) between the WT and KO condition. (D) Gene ontology analysis. The top five ontologies within biological process and cellular component are shown both for up- and down regulated proteins in METTL9 KO cells, relative to a WT control.