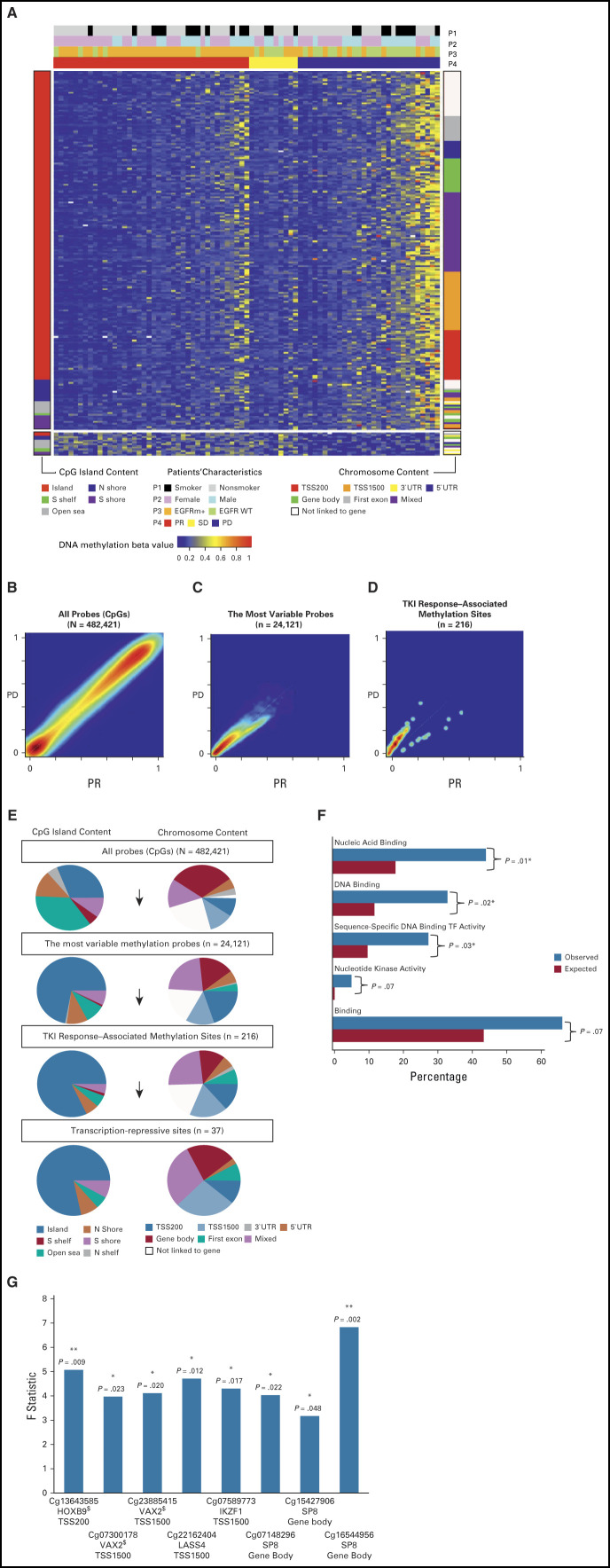
FIG 2.
DNA methylation landscape of 79 patients with lung adenocarcinoma with different EGFR-TKI responses. (A) DNA methylation heatmap of 216 EGFR-TKI response–associated sites for DCR prediction. The color-coded beta value at each selected probe ranged between 0 (blue) and 1 (red). The patients’ characteristics, smoking behavior (P1), sex (P2), EGFR status (P3), and EGFR-TKI response (P4), are shown by the bars at the top of the heatmap. Probes were grouped by CpG island content (left bar) and by chromosome content (right bar). (B-D) DNA methylation density for PD versus PR. For each CpG probe, the average beta value across patients with PD was plotted against that across patients with PR and is shown as a smooth kernel scatter plot. The set of probes used is indicated at the top of each plot, along with the number of probes. In the gradient scale, red represents the densest region, whereas purple represents the sparsest region; PR (red), SD (yellow), and PD (blue). (E) Genomic context distributions of CpG methylation sites. The probe distributions in the CpG context and the gene context are shown on the left and right, respectively, for all probes in the array (top), for the most variably methylated probes (upper middle), for the TKI response–associated methylation sites (lower middle), and for the transcription-repressive sites (bottom). (F) Functional enrichment analysis of genes linked to the 37 transcription-repressive sites. All 186 molecular function categories in GO-slim were evaluated, and the top 5 most enriched categories are shown, along with fold changes and P values. The GO terms involving transcription factors were GO:0003676, GO:0003677, and GO:0003700. (G) F value of ANOVA for the eight TF-linked sites. *P < .05; **P < .01; $enhancer (ENCODE). ANOVA, analysis of variance; DCR, disease control rate; EGFR, epidermal growth factor receptor; ENCODE, Encyclopedia of DNA Elements; GO, gene ontology; PD, progressive disease; PR, partial response; SD, stable disease; TF, transcription factor; TKI, tyrosine kinase inhibitor.