FIG 3.
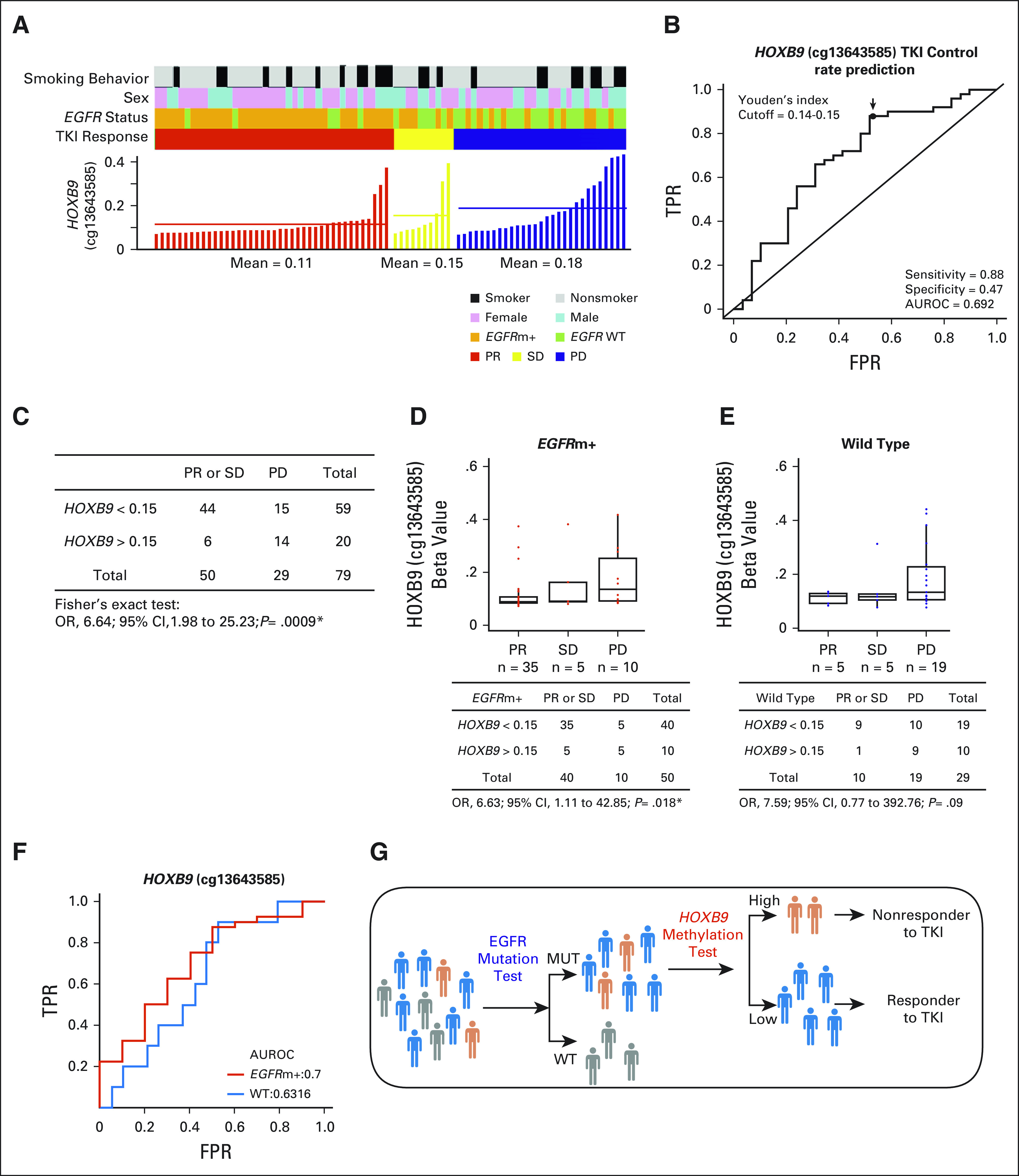
DNA methylation of HOXB9 (cg13643585) was correlated with EGFR-TKI response. (A) The HOXB9 beta values of 79 patients with NSCLC with PR (red), SD (yellow), and PD (blue). (B) AUROC and (C) Fisher’s test of HOXB9 methylation for predicting the EGFR-TKI response. . The optimal cutoff points were determined by the Youden's index, which maximizes the sum of the specificity and sensitivity. (D-F) Stratified analysis. Patients were classified as EGFRm+ (red) or EGFR wild type (blue). Comparison of HOXB9 beta value among PR, SD, and PD. (G) Strategy using HOXB9 methylation complementing EGFR to classify the subpopulation of patients likely to be nonresponders to EGFR-TKI. AUROC, area under the receiver operating characteristic curve; EGFR, epidermal growth factor receptor; FPR, false positive rate; NSCLC, non–small-cell lung cancer; OR, odds ratio; PD, progressive disease; PR, partial response; SD, stable disease; TKI, tyrosine kinase inhibitor; TPR, true positive rate; WT, wild type.
