Figure 7. Targeting KDM4A Overcomes SCC Resistance to PD1 Blockade and Eliminates CSCs by Recruiting CD8+ T cells into Tumors.
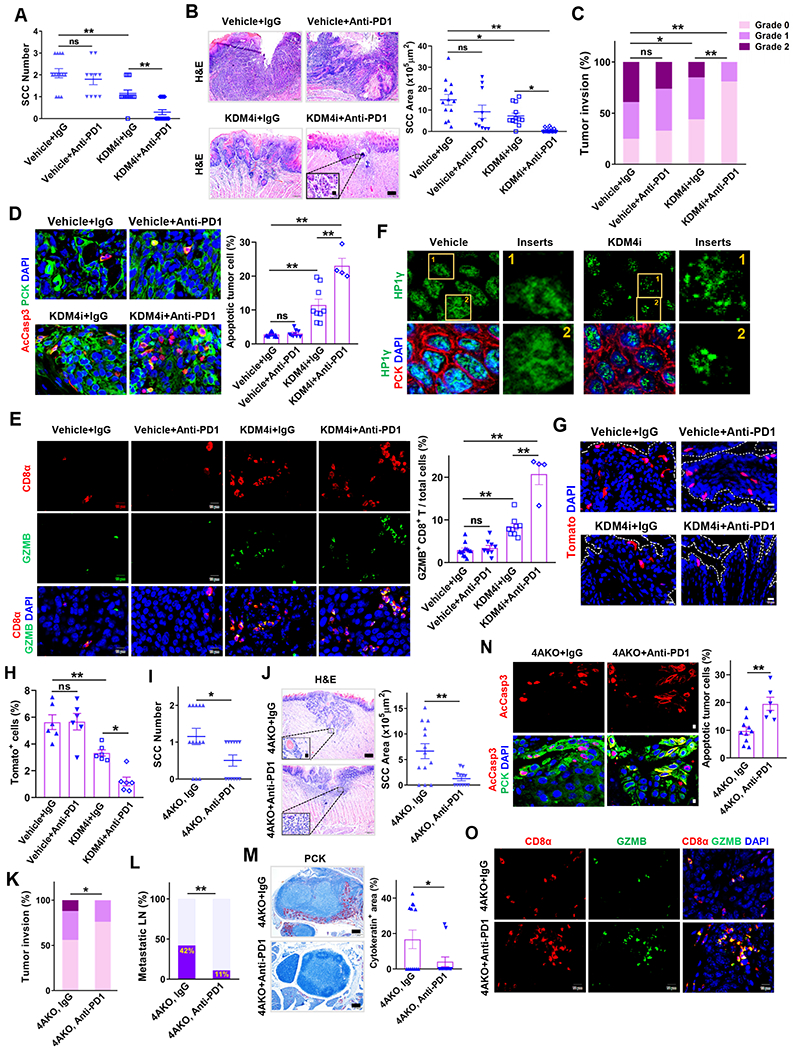
(A) Quantification of primary SCC number in mice treated with anti-PD1, KDM4i or their combination. Values represent mean ± SD from the pool of two independent experiments. n=13:10:13:17. ns, not significant, and **p < 0.01 by one-way ANOVA.
(B) H&E staining and area quantification of primary SCCs. Scale bar, 200 μm. Enlarged images were shown in inserts with 10 μm scale bar. n = 13:10:13:17. ns, not significant, *p < 0.05 and **p<0.01 by one-way ANOVA.
(C) Histogram analysis of primary tumor invasion grades. ns, not significant, *p<0.05 and **p<0.01 by Cochran-Armitage test.
(D) Immunofluorescence staining and quantification of apoptotic tumor cells. Values are mean ± SEM from the pool of two-independent experiments. n = 12:9:9:4. ns, and **p < 0.01 by one-way ANOVA.
(E) Immunofluorescence staining and quantification of GZMB+CD8+ T cells within and around tumor islets. Values represent mean ± SEM from the pool of two independent experiments. Scale bars, 10 μm. n = 12:9:9:4. ns, not significant, and **p<0.01 by one-way ANOVA.
(F) Representative immunofluorescence staining of HP1γ (green), PCK (red) and DAPI (blue) in primary SCCs of mice. Scale bar, 2 μm. Enlarged images were shown in inserts with 1 μm scale bar.
(G and H) Representative fluorescence images (G) and quantification (H) of tdTomato+ cells (BMI1+ CSCs) in primary SCCs of mice. n = 6:6:6:6. Scale bar 10 μm. ns, not significant, *p<0.05 and **p<0.01 by one-way ANOVA.
(I) Quantification of primary tongue SCC number in 4AKO mice treated with anti-PD1 or IgG. Values are mean ± SD from the pool of two independent experiments, n = 13:12. *p < 0.05 by Student’s t test.
(J) H&E staining and area quantification of primary SCCs. Scale bar for the main image, 200 μm. Inserts are enlarged images with 10 μm scale bar. Values are mean ± SD from the pool of two independent experiments, n = 13:12. **p < 0.01 by Student’s t test.
(K) Quantification of primary tumor invasion degree in 4AKO mice treated with anti-PD1 or IgG. n = 13:12. *p < 0.05 by Cochran-Armitage test.
(L) Percentage of metastatic lymph nodes in 4AKO mice treated with anti-PD1 or IgG. Data are collected from two independent experiments. n = 13:12. **p < 0.01 by Chisquare test.
(M) PCK immunostaining and quantification of metastatic areas in lymph nodes from 4AKO mice. n = 13:12. Values represent mean values ± SEM from two independent experiments. *p < 0.05 by Student’s t test. Scale bars, 200 μm.
(N) Immunofluorescence staining and quantification of apoptotic tumor cells in primary mouse SCCs in 4AKO mice treated by either anti-PD1 (n = 6) or IgG (n = 10). Values are mean ± SEM from the pool of two independent experiments. Scale bars, 10μm. **p<0.01 by Student’s t test.
(O) Immunofluorescence staining of GZMB+CD8+ T cells within and around tumor islets. Scale bars, 10 μm.
Also see Figures S6 and S7.
