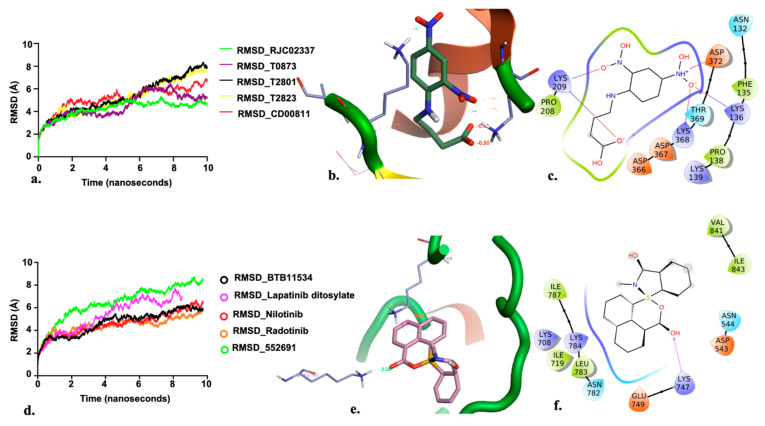
Figure 5.
(a) RMSD for the top five PfDegP-complexes from the allosteric site and it was quite evident that the two complexes RJC02337 and T0873 attained stability. The complex PfDegP-RJC02337 (green) was quite stable for the whole simulation period exhibiting the least fluctuations, whereas complex PfDegP-T0873 (purple) tends to attain its stability after a simulation period of ~8 nanoseconds. However, the complexes PfDegP-T2823 (black), PfDegP-T2801 (yellow) and CD00811 (red) exhibited a continuous increase in their RMSD, indicating their unstable interaction with the protein. (b) Least binding energy pose of Maybridge_RJC02337 docked with PfDegP (c) Ligand interaction diagram of Maybridge_RJC02337 showing the interacting residues with PfDegP (d) The RMSD for the top five PfDegP- complexes and it was quite evident that the DegP complex with BTB11534, Lapatinib ditosylate, Radotinib, 552691 attained stability while the complex with Nilotinib exhibited the increased RMSD (e) Least binding energy pose of Maybridge_BTB11534 docked with PfDegP (f) Ligand interaction diagram of Maybridge_BTB11534 showing the interacting residues with PfDegP.