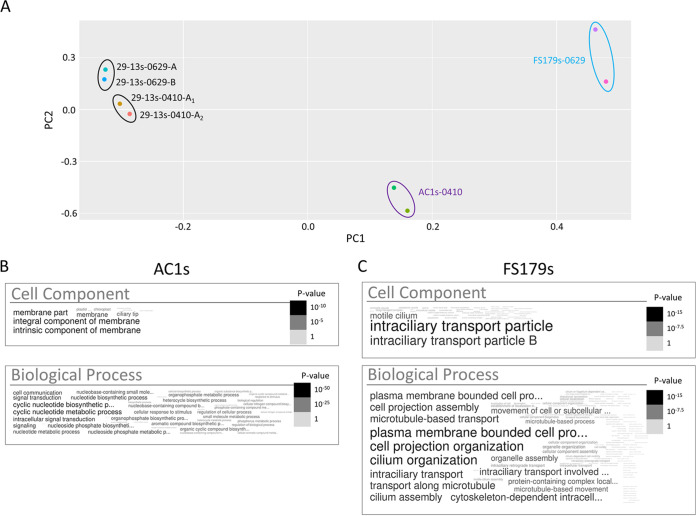
FIG 6.
APEX2 proximity proteomics differentiates protein compositions of flagellum subdomains. (A) Principal-component analysis of proteins identified in supernatant fractions from 29-13 (29-13s), AC1-APEX2 (AC1s), and FS179-APEX2 (FS179s) cells. For parental cell line controls (29-13), three independent experiments are shown (0410, 0629-A, and 0629-B), and for one experiment, the sample was split into two aliquots (0410-A1 and 0410-A2) that were subjected to shotgun proteomics in parallel. For FS179s, two independent experiments are shown (0629-A and 0629-B). For AC1s, one sample was split into two aliquots that were subjected to shotgun proteomics in parallel (0410-A1 and 0410-A2). (B) Word cloud representing GO analysis for cellular components and biological processes of the proteins identified in the AC1s proximity proteome. (C) Word cloud representing GO analysis for cellular components and biological processes of the proteins identified in the FS179s proximity proteome.