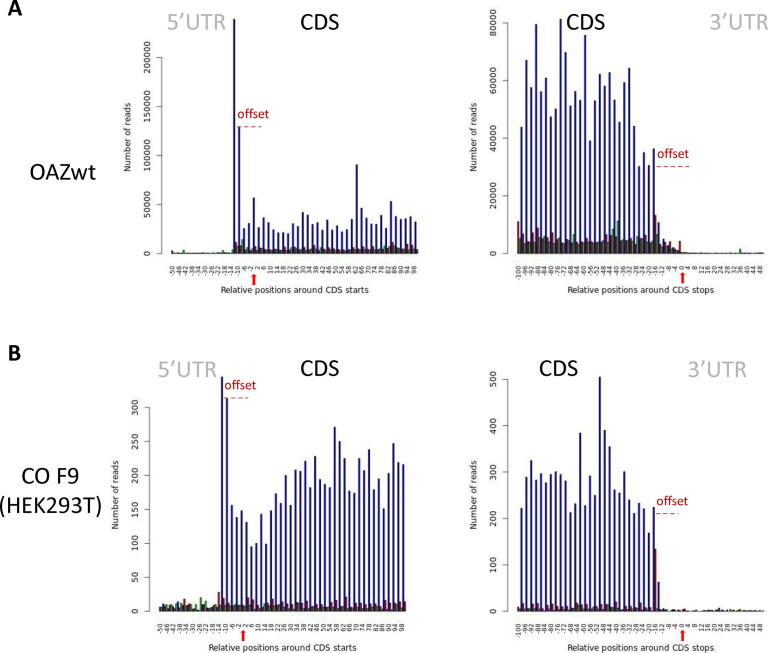
Fig. 3.
Metagene periodicity. Each graph shows the coverage of the CDS and UTR regions of the metagene for a specific read length (here 28 nucleotides), determined by representing all the reads starting at a specific relative position. Each of the three possible reading frames is represented in a different color (blue, green and red). The coverage is shown according to the window chosen by the user. In this example: from 50 nucleotides before the start codon to 100 nucleotides after the start codon, and then from 100 nucleotides before the stop codon to 50 nucleotides after the stop codon. To help the reader, we manually added the red arrows to indicate the position of the START and STOP codons and the offset observed that is due to only the first nucleotide on the 5′ side of the reads being counted. A) Periodicity for the OAZwt yeast strain. B) Periodicity for the HEK293T cell line. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)