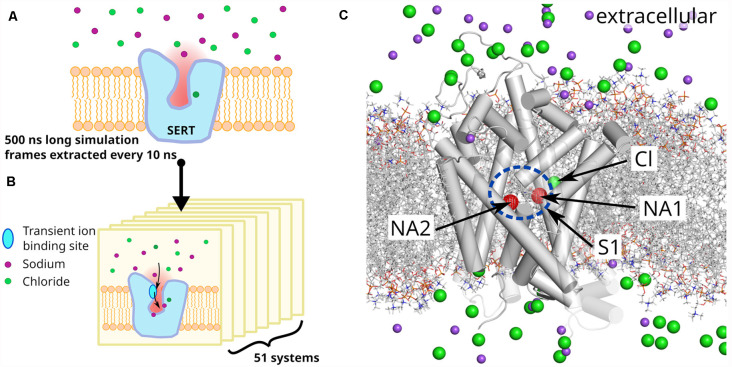
Figure 1.
System and system setup. (A) A 500 ns long unbiased simulation of serotonin transporter (SERT) in the absence of bound sodium ions was carried out to prepare a set of starting systems. (B) Fifty-one snapshots separated by 10 ns were extracted and sodium ions removed, if present in the outer vestibule. All systems were independently simulated for 150 ns. (C) Representative starting structure of a selected SERT snapshot, showing sodium (purple) and chloride (green) ions as spheres and highlighting the sodium binding sites NA1 and NA2 by red spheres. The substrate binding site S1 is indicated by a blue ellipse. The lipid membrane is represented by gray dashed lines. NA1 is formed by TM1, TM6 and TM7, NA2 by TM1 and TM8. Temporary binding sites are formed by D326, E493, and E494 from which E493 is part of the extracellular gate (EC) with R104.