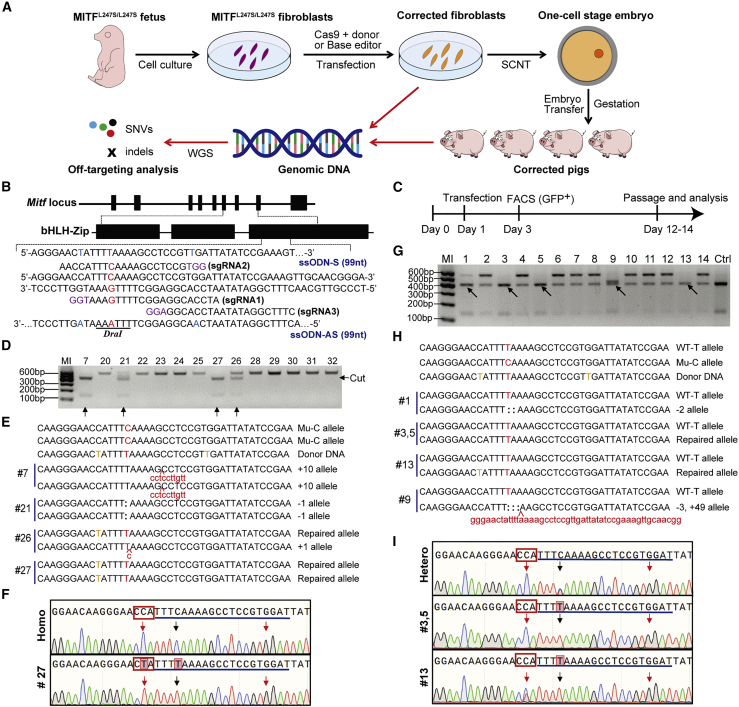
Figure 1.
CRISPR-Cas9 mediated MITF c.740C > T repair in porcine fibroblast cells using ssODN as the repair template
(A) Experimental design for the MITF point-mutation repair in the current study. (B) Genome structure of the porcine MITF gene, the sequences of the sgRNAs targeting the c.740 site, and the ssODNs used as HDR templates. The protospacer-adjacent motif (PAM) sequences of the sgRNAs are shown in purple, the intended point mutation is shown in red, and the introduced blocking mutations are shown in blue in the ssODN sequence. (C) Experimental procedure for screening single-cell colonies. (D) Genotyping of single-cell colonies by PCR and DraI digestion that originated from MITFL247S/L247S fibroblasts. The arrows indicate potentially corrected colonies. (E) Sanger sequencing of the colonies cut by DraI. The T base intended to be repaired is shown in red, the blocking mutations in donor DNA are shown in orange, the deleted bases are indicated by colons, and the inserted bases are shown by lowercase red text. (F) Sequencing diagram of the PCR product from colony 27 (#27) and the MITFL247S/L247S fibroblast cells (Homo). The sgRNA sequences are underlined, the PAM sequences of the sgRNA1 are shown in red rectangles, and the repaired base and the introduced blocking mutation are indicated by black and red arrows, respectively. (G) Genotyping of single-cell colonies by PCR and DraI digestion originate from MITFL247S/+ fibroblasts. The arrows indicate the potentially corrected colonies. (H) Sanger sequencing of the colonies cut by DraI. The T base intended to be repaired is shown in red, the blocking mutations in donor DNA are shown in orange, the deleted bases are indicated by colons, and the inserted bases are shown by lowercase red text. (I) Sequencing diagram of the PCR product from colony 3, 5, and 13 and the MITFL247S/+ fibroblast cells (Hetero). The PAM sequences of the sgRNA1 are shown in red rectangle, and the repaired base and the introduced blocking mutation are indicated by black and red arrows, respectively.