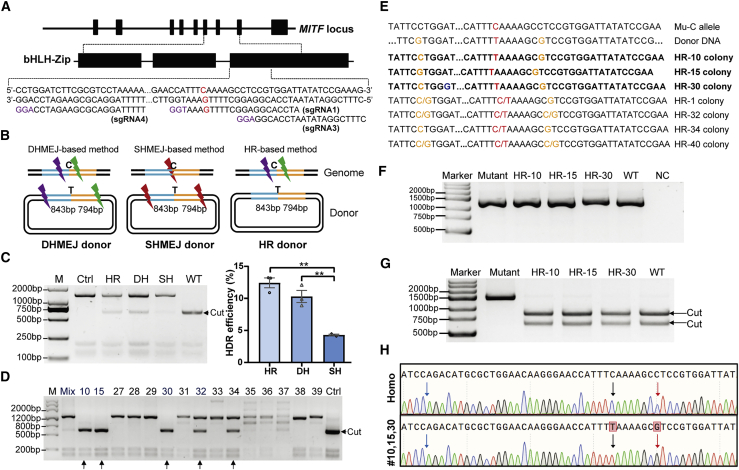
Figure 3.
CRISPR-Cas9-mediated MITF point mutation repair in MITFL247S/L247S porcine fibroblast cells using long donor plasmids as the repair templates
(A) Design of different sgRNAs around exon 8 of the porcine MITF gene for the long donor DNA strategy. The PAM sequences of sgRNAs are shown in purple, and the intended point mutation is shown in red. (B) Design of donor DNA and sgRNAs for genome editing. The homology arms are shown in blue and orange lines around the intended T locus, and the different sgRNAs are shown as lightning bolts. The purple lightning bolt represents sgRNA4, the green lightning bolt represents sgRNA3, and the red lightning bolt represents sgRNA1. (C) Analysis of the HDR efficiency for the three different schemes. A representative restriction fragment length polymorphism (RFLP) image of transfected cells is shown on the left, and the Hi-TOM quantitative results of three independent replicates are presented as mean ± SEM (n = 3) on the right. ∗∗p < 0.01. HR, DH, and SH represent HR, DHMEJ, and SHMEJ, respectively. (D) Genotyping of single-cell colonies by PCR and DraI digestion. The arrows indicate the potential repaired colonies. (E) Sanger sequencing of the colonies that were precisely repaired at the intended locus. The biallelically corrected colonies are shown in bold. The T base intended to be repaired is shown in red, the blocking mutations in the donor DNA are shown in orange, and the omitted bases are shown by ellipses. (F and G) Total RNAs were extracted from these colonies and amplified using RT-PCR (F) and then digested with DraI (G). Arrows indicate cleavage. (H) Sequencing diagram of the RT-PCR product of colony HR-10, HR-15, and HR-30 and the MITFL247S/L247S fibroblast cells (Homo). The intended corrected base and the introduced blocking mutation are indicated by arrows in black and red, respectively. The blue arrow indicates the junction of exon 7 and exon 8 of MITF.