Figure 5.
Correction of the MITF c.740T>C mutation using BEs
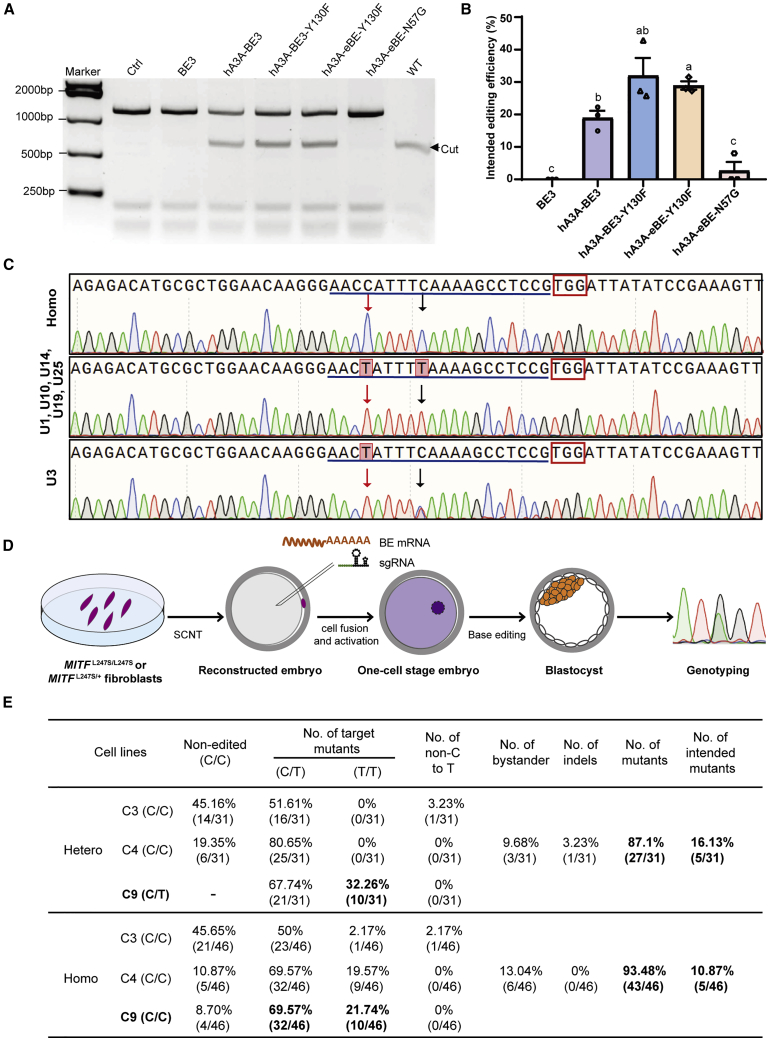
(A) RFLP analysis in MITFL247S/L247S fibroblast cells that were transfected with different BEs as indicated. (B) Hi-TOM was used to evaluate the editing efficiency of the intended correction with different BEs. The sequences with C9 corrected to T or both C9 and C4 mutated to T were counted as corrected events. Data are presented as mean ± SEM (n = 3 from three independent experiments). a, b, and c values with no letter in common are significantly different (p < 0.05). (C) Representative Sanger sequencing diagrams of the corrected colonies compared with the MITFL247S/L247S cells (Homo). The sgRNA sequences are underlined, and the PAM sequences are shown in red rectangles. The edited bases are indicated by arrows, the black arrows indicate the intended mutations (C9), and the red arrows indicate the C4 mutations in the editing window. (D) Schematic of microinjection in MITFL247S/L247S and MITFL247S/+ fibroblast-derived embryos. (E) Summary of the editing efficiency in heterozygous (hetero) and homozygous (homo) embryos by microinjection of sgRNA and hA3A-eBE-Y130F mRNA. Sequences with C9 corrected to T/T and C3 unedited were counted as intended mutants.