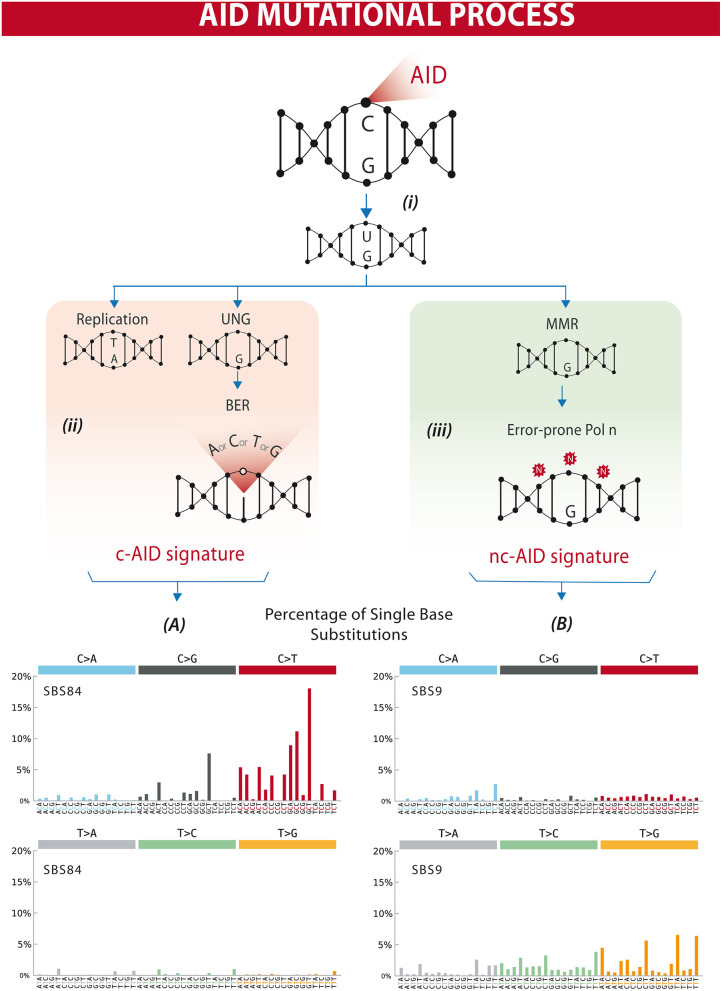
Figure 1.
Differential processing of AID lesions. (i) AID deaminates cytosine residues on single-stranded DNA that is exposed during transcription, converting C (cytosine) into U (uracil). (ii) The U-G (guanine) mismatch can be processed through different pathways. Either by replication that will result in a C>T transition or by uracil DNA glycosylase (UNG) followed by base excision repair (BER) resulting in C>T/G/A substitutions or homologous repair. This will often result in a mutational profile known as canonical AID signature (c-AID). (iii) On the other hand, mismatch-repair proteins (MMR) can also recognize and process AID-induced lesions. Exonucleases resect the abasic sites, which are followed by error-prone polymerase repair. This processing often results in a mutational profile similar to the non-canonical AID signature. Lower panels (A,B) depict mutational profiles using the conventional 96 mutation type classification. This classification is based on the six substitution subtypes: C>A, C>G, C>T, T>A, T>C, and T>G (all substitutions are referred to by the pyrimidine of the mutated Watson–Crick base pair). Each of the substitutions is examined by incorporating information on the bases immediately 5′ and 3′ to each mutated base generating 96 possible mutation types (6 types of substitution × 4 types of 5′ base × 4 types of 3′ base). Mutational signatures are displayed and reported based on the observed trinucleotide frequency of the human genome, i.e., representing the relative proportions of mutations generated by each signature based on the actual trinucleotide frequencies of the reference human genome version GRCh37. (A) SBS84 is found in clustered mutations in the immunoglobulin gene and other regions in lymphoid cancers. (B) SBS9 may be due in part to mutations induced during replication by polymerase eta. Mutation frequencies were retrieved from the Comic Catalog v3.1 (cancer.sanger.ac.uk) (13).