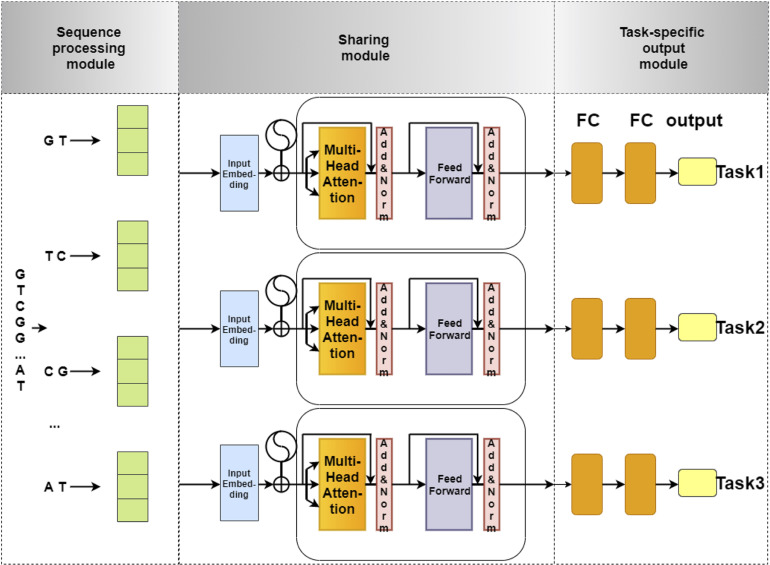
FIGURE 1.
The flowchart of 4mCPred-MTL. The sequence processing module uses 2-gram to split an original DNA sequence into overlapping subsequences and converts them into feature vectors by one-hot encoding. Next, the feature vectors of subsequences are fed into the sharing module, containing a Transformer encoder and a max-pooling layer, to capture the sharing information among different species. Finally, the output of the sharing module is fed into the task-specific output module to predict the 4mC site of a certain species.