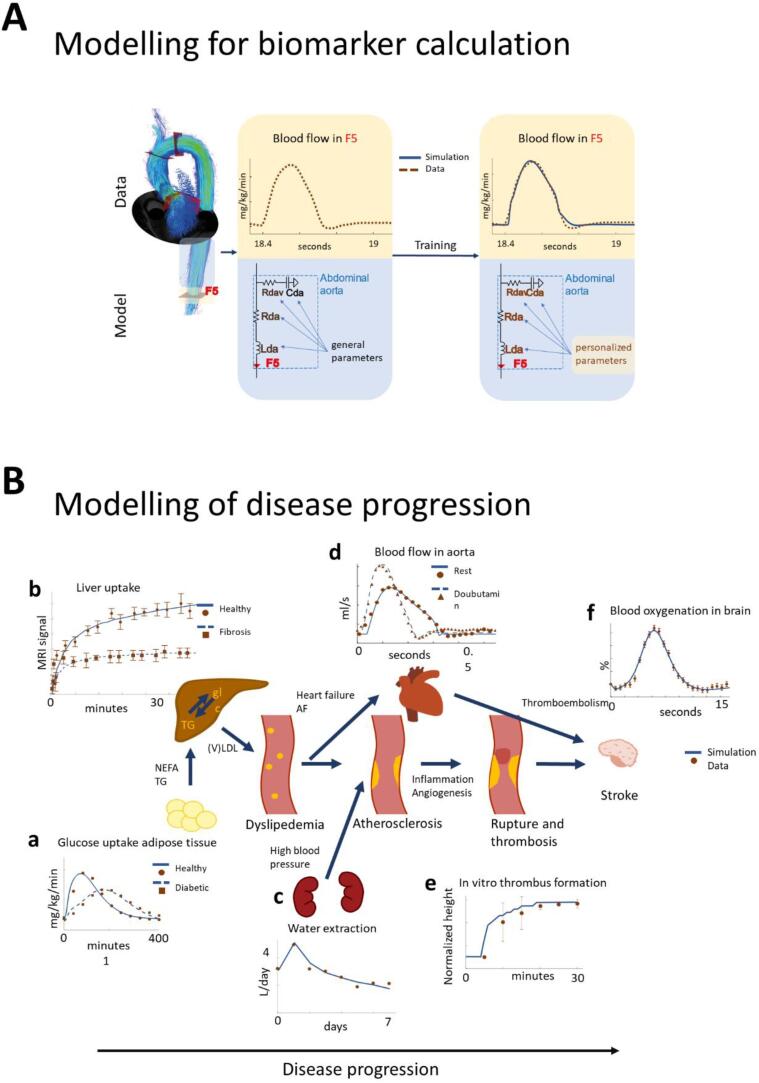
Fig. 5.
Abilities of mechanistic models. (A) The basic principle by which biomarkers can be calculated from data using mechanistic models, here illustrated by modelling of vessels around the aorta using a standard Windkessel model and 4D flow MRI data (Casas et al., 2017, Casas et al., 2018). Here, only a part of the model is shown, the one describing blood flow in the abdominal aorta. The general parameters are taken from literature. The personalized parameters received by training of the entire model can then be interpreted as personalized values of biomarkers. In the illustration, these biomarkers are the resistance (Rda), inertance (Lda), and capacitance (Cda) of the abdominal aorta, and the viscoelastic resistance (Rdav) of Cda. All of these are mechanistic and interpretable biomarkers, not available in the raw data, but whose values can be estimated using the mechanistic model. (B) Overview of some of the most important sub-models that could form the basis of a stroke simulation model, with agreements between simulations (-,--) and data (dots) in most of the main processes leading up to a stroke. (a) A model for adipose tissue meal response (at t = 0) in healthy controls (-) and type 2 diabetes patients (--)12; (b) Liver models describing liver uptake of a contrast agent injected at t = 0, in patients ranging from healthy controls-) to advanced fibrosis--)94; (c) The kidney water extraction response following administration of an SGLT2 inhibition drug at t = 095; (d) Blood flow in aorta for normal healthy subjects (-) and for subjects perturbed with Dobutamine (--)10; (e) Thrombus formation in an in vitro vessel initiated at t = 096; (f) The Blood Oxygen Level Dependent signal in response to brain activity (Sten et al., 2017). These models are currently isolated from each other, but could become a part of an interconnected stroke simulation model. Such interconnected simulations should still be different from patient to patient.