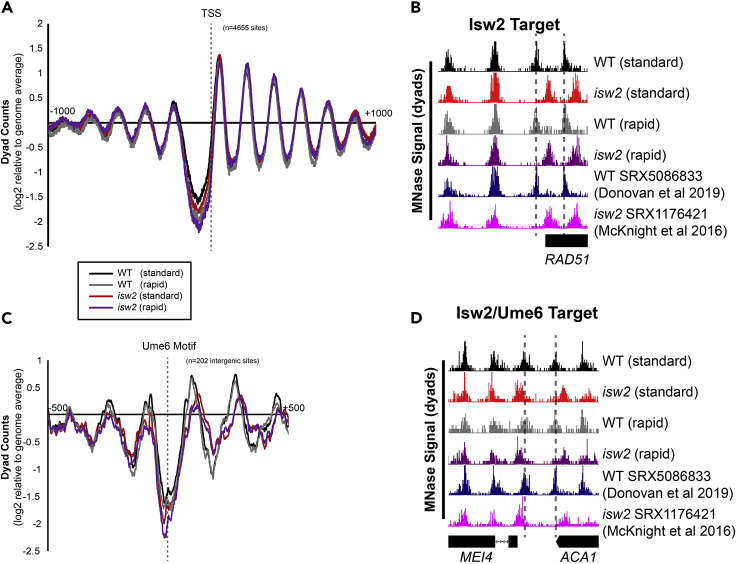
Figure 2.
Rapid MNase can accurately map nucleosome positions in S. cerevisiae cells
(A) Nucleosome dyad signal at 4655 yeast transcription start sites (TSSs) are plotted for WT and isw2 nucleosomes harvested using a standard or rapid protocol.
(B) Example Genome Browser image showing the standard method and rapid method can map Isw2-directed nucleosome positions similar to published data sets at the RAD51 locus. Dashed lines indicate Isw2-positioned nucleosomes.
(C) Nucleosome dyad signal at 202 intergenic Ume6 target sites showing rapid and standard MNase methods can accurately identify global changes in nucleosome structure at an Isw2 recruitment motif.
(D) Genome Browser image showing all methods correctly identify Isw2-positioned nucleosomes at the MEI4-ACA1 locus, a Ume6 target site. Dashed lines indicate Ume6- and Isw2- positioned nucleosomes.