Figure 3.
Rapid MNase can recover nucleosome footprints from isolated quiescent cells and yeast patches
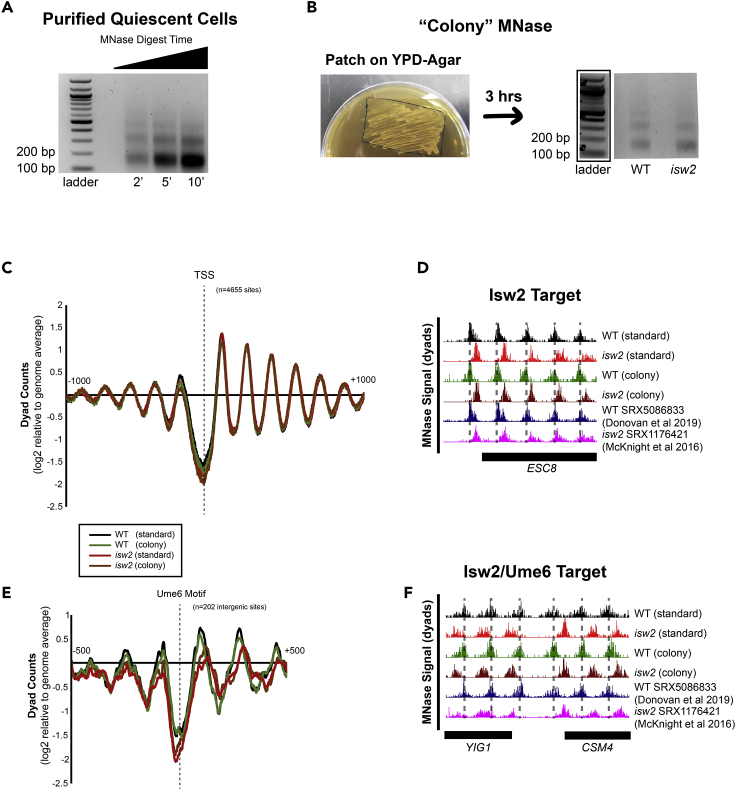
(A) Representative gel showing nucleosome footprints recovered from purified quiescent cells using the rapid MNase protocol.
(B) Representative gel (right) showing nucleosome footprints recovered from a fresh patch of yeast collected from YPD-Agar (left).
(C) Nucleosome dyad signal at transcription start sites (TSSs) comparing standard and patch- recovered “colony” MNase footprints.
(D) Example Genome Browser image showing “colony” MNase footprints can accurately identify Isw2-directed nucleosome positions at the ESC8 locus compared to the standard MNase protocol and published data sets.
(E) Nucleosome dyad signal at 202 intergenic Ume6 target sites showing “colony” MNase can accurately identify global changes at an Isw2 recruitment motif.
(F) Genome Browser image showing colony MNase can similarly identify Isw2-positioned nucleosomes at the YIG1-CSM4 locus, an example Ume6 target site. Dashed lines indicate Ume6- and Isw2-positioned nucleosomes.