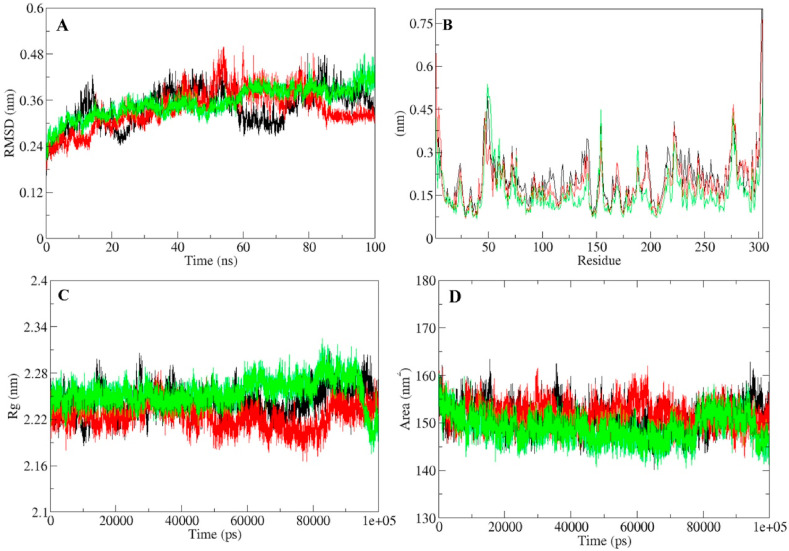
Fig. 4.
Molecular dynamic simulation trajectory analysis of SARS-CoV-2 Mpro protein and protein-ligand complexes during 100 ns simulation (A) RMSD of solvated SARS-CoV-2 Mpro protein, Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex during 100 ns MD simulation (B) RMSF values of Mpro protein, Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex (C) Rg during 100 ns MD simulation of Mpro protein, Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex. (D) SASA during 100 ns MD simulation of Mpro protein, Mpro-lopinavir and Mpro-quercetin-3-rutinoside-7-glucoside complex. Unbound protein parameters are depicted in black color. Parameters for Mpro-lopinavir complex and Mpro-quercetin-3-rutinoside-7-glucoside complex are represented in red and green color respectively.