Abstract
Dental caries is characterized by a dysbiotic shift at the biofilm–tooth surface interface, yet comprehensive biochemical characterizations of the biofilm are scant. We used metabolomics to identify biochemical features of the supragingival biofilm associated with early childhood caries (ECC) prevalence and severity. The study’s analytical sample comprised 289 children ages 3 to 5 (51% with ECC) who attended public preschools in North Carolina and were enrolled in a community-based cross-sectional study of early childhood oral health. Clinical examinations were conducted by calibrated examiners in community locations using International Caries Detection and Classification System (ICDAS) criteria. Supragingival plaque collected from the facial/buccal surfaces of all primary teeth in the upper-left quadrant was analyzed using ultra-performance liquid chromatography–tandem mass spectrometry. Associations between individual metabolites and 18 clinical traits (based on different ECC definitions and sets of tooth surfaces) were quantified using Brownian distance correlations (dCor) and linear regression modeling of log2-transformed values, applying a false discovery rate multiple testing correction. A tree-based pipeline optimization tool (TPOT)–machine learning process was used to identify the best-fitting ECC classification metabolite model. There were 503 named metabolites identified, including microbial, host, and exogenous biochemicals. Most significant ECC-metabolite associations were positive (i.e., upregulations/enrichments). The localized ECC case definition (ICDAS ≥1 caries experience within the surfaces from which plaque was collected) had the strongest correlation with the metabolome (dCor P = 8 × 10−3). Sixteen metabolites were significantly associated with ECC after multiple testing correction, including fucose (P = 3.0 × 10−6) and N-acetylneuraminate (p = 6.8 × 10−6) with higher ECC prevalence, as well as catechin (P = 4.7 × 10−6) and epicatechin (P = 2.9 × 10−6) with lower. Catechin, epicatechin, imidazole propionate, fucose, 9,10-DiHOME, and N-acetylneuraminate were among the top 15 metabolites in terms of ECC classification importance in the automated TPOT model. These supragingival biofilm metabolite findings provide novel insights in ECC biology and can serve as the basis for the development of measures of disease activity or risk assessment.
Keywords: children, biofilm, dental caries, microbiome, machine learning, risk assessment
Introduction
Early childhood caries (ECC) is a persistent clinical and public health problem with multilevel consequences (Casamassimo et al. 2009). The prevalence of untreated caries has almost halved during the past decade, but more than 1 in 3 US children are still affected by ECC. The disease is influenced by social, environmental, and behavioral factors, but fermentable carbohydrate consumption and inadequate fluoride exposure are its most proximal and well-studied risk factors (Pitts et al. 2017). The pathogenesis of dental caries occurs at the biofilm–tooth surface interface as a result of a dysbiotic, cariogenic microbial community that interacts with environmental and host factors (Nyvad et al. 2013; Bowen et al. 2018). Despite notable advances in the basic and clinical sciences, comprehensive characterization of the molecular and biochemical profile of the ECC-associated microbial imbalance and virulence remains elusive.
Understanding the biological (i.e., microbial, biochemical, environmental) basis of ECC is arguably one of the missing keys needed for the development of effective diagnostic, preventive, and disease management approaches (Divaris 2016). Recent next-generation sequencing studies offer characterizations of the oral microbiome in higher resolutions than ever before (Nascimento et al. 2017; Mira 2018). Previously unrecognized bacterial species, important for health and disease, have emerged (Hajishengallis et al. 2017; Rosier et al. 2018; Hurley et al. 2019). Interestingly, recent evidence indicates that Candida (Klinke et al. 2009; Xiao et al. 2018), viruses (Yildirim et al. 2010), and other nonbacterial organisms may be important activists in the supragingival biofilm of children with ECC.
While important new information regarding the composition of the ECC-associated biofilm is emerging, little is known about its biochemicals and their functional activity. The metabolic profile of dental plaque is arguably “where the rubber meets the road” for the pathogenesis of dental caries. One study to date has examined childhood caries-associated metabolites in the supragingival biofilm among 11 caries-active and 4 caries-free children between ages 10 and 15 y (Zandona et al. 2015). Those results indicated the existence of biofilm metabolites with the potential to provide a metabolomics signature for caries activity. We embarked upon the present study with the overarching goal of addressing the knowledge gap in biofilm metabolomics for ECC. Specifically, we sought to identify biochemical metabolites in the supragingival biofilm that are associated with ECC prevalence and severity.
Methods
Study Population and Characterization
The initial study population comprised 300 preschool-age children attending public preschool (Head Start) centers in North Carolina, participants of a community-based, cross-sectional, epidemiologic study (ZOE 2.0) of early childhood oral health (Divaris and Joshi 2020; Divaris et al. 2020). Children were between 36 and 71 mo old. Their legal guardians provided written informed consent for clinical data and biospecimen (saliva and dental plaque) collection. The study received ethics approval by the University of North Carolina (UNC)–Chapel Hill Institutional Review Board (#14-1992). Children underwent comprehensive dental examinations recording dental caries experience using International Caries Detection and Classification System (ICDAS) criteria by trained and calibrated examiners between August 2016 and February 2019. Detailed descriptions of the clinical examination (Ginnis et al. 2019), biofilm collection, and analysis protocols (Divaris et al. 2019), as well as the parent study’s cohort profile (Divaris et al. 2020), have been reported in detail in previous publications.
Supragingival Biofilm Sample Collection
The supragingival biofilm samples were collected prior to the dental examinations, which took place before or at least 30 min after breakfast or snack. The families were instructed not to brush their child’s teeth the morning of the clinical encounter. A plaque sample for metabolomics analyses was collected from the facial/buccal surfaces of the upper-left quadrant (Universal system: #F, #G, #H, #I, and #J; FDI system: #61, #62, #63, #64, and #65). Examiners used sterile toothpicks for plaque collection, and the samples were immediately frozen at the collection site (−20°C using CoolBoxes, BCS-575, Brooks Life Sciences) until being transferred to a university core facility and stored long term at −80°C until further processing.
Metabolomics Analyses
The metabolomic analysis was done by Metabolon using the proprietary DiscoveryHD4 (Metabolon Inc) platform (Evans et al. 2009, 2014) that includes multiple mass spectrometry methods, a large reference library of authenticated metabolite standards, and a suite of patented informatics and quality control software. This global metabolomics methodology allows for the detection of metabolites in all major metabolite classes. The ultra-performance liquid chromatography–tandem mass spectrometry pipeline enables the identification of biochemical metabolites in the plaque samples by comparison to library entries of purified standards or recurrent unknown entities, using metrics including the retention time/index, mass-to-charge ratio, and chromatographic database criteria on all molecules in the library maintained by Metabolon. Detailed descriptions of the metabolomics data generation procedures, including laboratory, informatics, normalization, and quality control procedures, have been previously reported (Divaris et al. 2019).
Clinical Comparison Groups
We initially selected 300 study participants for this nested case-control metabolomics study, aiming to maximize power: the first 150 presenting as “established” ECC cases (i.e., had at least 1 restored or missing surface due to caries or caries lesions detected at the ICDAS ≥3 threshold) and 150 participants who did not meet this criterion. Subsequently, 1 participant was excluded from analyses because, although eligible at study enrollment, he or she was 73 mo old at the time of clinical examination and thus outside the age range of ECC definition. Ten additional participants (3 with ECC defined at the ICDAS ≥3 threshold and 7 without) were excluded from analyses based on a high proportion of missing individual metabolite data (>30%) (Appendix Fig. 1) and clustering close to blank control specimens during quality control procedures.
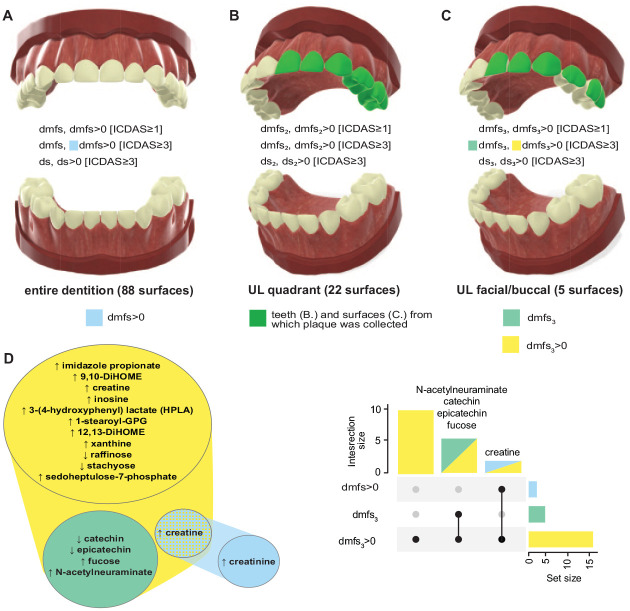
We defined and examined 18 traits of ECC prevalence and severity according to 3 definitions: caries experience including early stage lesions detected at the ICDAS ≥1 threshold or classic ECC (Pitts et al. 2019), caries experience including established/advanced caries lesions detected at ICDAS ≥3 threshold, and untreated disease only defined as lesions at the ICDAS ≥3 threshold. Traits were defined within 3 sets of tooth surfaces, i.e., the entire dentition (88 surfaces), all surfaces of the sampled teeth (22 surfaces), and the 5 facial/buccal surfaces from which plaque was collected (Fig. 1). ECC was measured using a continuous measure of disease experience (i.e., the sum of decayed, restored, or missing tooth surfaces [dmfs index] for each definition and tooth set) and a corresponding binary case definition (dmfs >0 for caries experience, ds >0 for untreated disease). ECC measures defined within the 22 surfaces of the sampled teeth are presented with subscript-2 indicators (i.e., dmfs2), and those defined within the 5 specifically sampled surfaces (i.e., the localized traits) are presented with subscript-3 indicators (i.e., dmfs3).
Figure 1.
Visual representations of the early childhood caries (ECC) traits that were defined and used in metabolomics analyses according to 3 different sets of tooth surfaces and summary of the significantly associated metabolites. (A) Entire dentition (88 surfaces, continuous and binary traits for 2 International Caries Detection and Classification System [ICDAS] thresholds and untreated disease). (B) All surfaces of teeth from which biofilm was sampled (22 surfaces), green highlighted. (C) The specific 5 facial/buccal surfaces from which biofilm was sampled, green highlighted. (D) Venn diagram and UpSet plot of metabolites significantly associated with more than 1 dental caries trait (linear regression of log2-transformed metabolite values with false discovery rate correction, q < 0.05). Arrows indicate the direction of the association. Catechin, epicatechin, fucose, and N-acetylneuraminate were associated with 2 localized ICDAS ≥1 disease traits, and creatine was associated with a localized binary ICDAS ≥1 trait and the ECC person-level case definition at the ICDAS ≥3 detection threshold.
Analytical Approach
We used feature-wise quantile regression imputation of left-censored data (QRILC) (Wei et al. 2018) to impute missing metabolite data. We examined the correlation of ECC traits and candidate covariates with the entire metabolome using Brownian distance correlations (dCor) (Székely and Rizzo 2009) and corresponding P values that were obtained with a permutation bootstrap (999 replicates). Upon examination of metabolome correlations with children’s age (measured in months), gender (male, female), and race/ethnicity (non-Hispanic White [NHW], African American [non-Hispanic Black, NHB], other), we found significant associations with age (dCor = 0.233, P = 3 × 10−2) and race/ethnicity (dCor = 0.234, P = 1 × 10−3) but not gender. Therefore, individual metabolite associations were examined using crude and age- and race/ethnicity-adjusted estimates from linear regression modeling of log2-transformed values. A false discovery rate (FDR) multiple testing correction (Benjamini and Hochberg 1995) and a 5% significance level were used to identify statistically significantly altered metabolites.
We used an automated machine learning (ML) approach to identify the best ECC classification model using all information contained in the metabolome (i.e., all 503 metabolites) and determine the discriminatory ability of the significantly altered metabolites in this context. For this application, we prioritized the binary ECC case definition with the highest Brownian distance correlation with the metabolome. We used a tree-based pipeline optimization tool (TPOT)–based automated algorithm (Olson and Moore 2019) that employs genetic programming to build pipelines of ML methods for classification, along with preprocessing operators such as data transformers and feature selectors. In the algorithm optimization phase, various combinations of transformers are combined with ML methods into a pipeline in a tree-based manner, and the best-performing pipeline is selected at the end of that process. Fifty random seed TPOT replicates were run for a maximum of 24 h, using 10-fold cross-validation with balanced accuracy (i.e., unweighted average of the number of correct predictions from all predictions calculated on per-class basis) as the performance estimate. The TPOT-identified model performance was compared against grid search–optimized logistic regression, random forest, and gradient boosting classifiers. Once the model was identified, we estimated individual metabolites’ predictive ability or “feature importance” coefficients and corresponding rank orders using a permutation feature importance (PFI) approach (Breiman 2001), as in a recent metabolomics application (Orlenko et al. 2020). Analyses and visualizations were done using R and Python packages and Stata 16.1 (StataCorp LP). Reporting of this observational study conforms with the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines (von Elm et al. 2007). The log2-transformed QRILC imputed data set used for the present analysis alongside metabolite biochemical information is available in the Appendix. The raw metabolomics data have been deposited to the EMBL-EBI MetaboLights database (Haug et al. 2020) with the identifier MTBLS2215. The data set can be accessed at https://www.ebi.ac.uk/metabolights/MTBLS2215.
Results
Study participants had a mean age of 52 mo (range, 36–71), and 53% were male and of mixed racial/ethnic distribution: NHB (37%), NHW (30%), and others, including Hispanics and those with more than 1 race (32%). The distribution of the 18 ECC clinical traits is presented in Table 1—noteworthy, most participants (95%) had ECC using the classic case definition (corresponding dmfs mean = 14), half of them (51%) had ECC according to the established/severe (ICDAS ≥3) definition (corresponding dmfs mean = 7), and a third (34%) had at least 1 untreated caries lesion (corresponding ds = 1.7). These estimates were lower but still appreciable when considering subsets of tooth surfaces; for example, 39% of participants had classic ECC when considering its localized definition (dmfs3 >0).
Table 1.
Distribution Characteristics of the Examined Clinical Traits of ECC Experience and Their Associations with the Metabolome and Significantly Altered Individual Metabolites.
| Characteristic | dmfs >0 (ICDAS ≥1), Proportion (SD) | dmfs (ICDAS ≥1), Mean (SD); Median (Range) | dmfs >0 (ICDAS ≥3), Proportion (SD) | dmfs (ICDAS ≥3), Mean (SD); Median (Range) | ds >0 (ICDAS ≥3), Proportion (SD) | ds (ICDAS ≥3), Mean (SD); Median (Range) |
|---|---|---|---|---|---|---|
| Descriptive information of the 18 examined ECC traits | ||||||
| Entire dentition (88 surfaces) | 0.95 (0.22) | 14.23 (15.61); 8 (0, 74) | 0.51 (0.50) | 7.03 (13.76); 1 (0, 69) | 0.34 (0.48) | 1.66 (4.03); 0 (0, 30) |
| All surfaces of the upper-left quadrant teeth (22 surfaces) | 0.84 (0.37) | 4.08 (4.90); 2 (0, 22) | 0.38 (0.49) | 2.12 (4.46); 0 (0, 22) | 0.22 (0.41) | 0.52 (1.64); 0 (0, 14) |
| All facial/buccal surfaces of the upper-left quadrant teeth (5 surfaces) | 0.39 (0.49) | 0.95 (1.45); 0 (0, 5) | 0.2 (0.4) | 0.45 (1.08); 0 (0, 5) | 0.07 (0.25) | 0.10 (0.44); 0 (0, 3) |
| Estimates of association with the entire metabolome and numbers of individual positively and negatively associated metabolites for the 18 examined ECC traits | ||||||
| Entire dentition (88 surfaces) | ↑0.150 (0.799) ↑1 ↓8 | ↑0.216 (0.164) ↑14 ↓12 | ↑0.192 (0.021) ↑51 ↓7 | ↑0.204 (0.256) ↑2 ↓4 | ↑0.194 (0.029) ↑36 ↓50 | ↑0.230 (0.020) ↑28 ↓7 |
| All surfaces of the upper-left quadrant teeth (22 surfaces) | ↑0.156 (0.559) ↑5 ↓7 | ↑0.206 (0.354) ↑11 ↓12 | ↑0.184 (0.046) ↑56 ↓1 | ↑0.192 (0.471) ↑2 ↓6 | ↑0.198 (0.014) ↑69 ↓2 | ↑0.226 (0.028) ↑17 ↓14 |
| All facial/buccal surfaces of the upper-left quadrant teeth (5 surfaces) | ↑0.208 (0.008)↑52 ↓14 | ↑0.220 (0.019) ↑22 ↓11 | ↑0.172 (0.142) ↑38 ↓2 | ↑0.189 (0.277) ↑5 ↓7 | ↑0.201 (0.013) ↑80 ↓9 | ↑0.212 (0.010) ↑41 ↓16 |
indicates Brownian distance correlation coefficient (P value). ↑ indicates number of positively associated metabolites, and ↓ indicates number of negatively associated metabolites, at a nominal significance level (P < 0.05 without multiple testing correction), derived from a linear regression model of log2-transformed metabolite values adjusting for participants’ age and race/ethnicity.
dmfs, the sum of decayed, missing, and restored (i.e., “filled”) primary tooth surfaces due to caries; ds, the sum of decayed primary tooth surfaces (i.e., “unrestored disease”); ECC, early childhood caries; ICDAS, International Caries Detection and Classification System (1: first visual change in enamel [“initial lesion”]; 3: localized enamel breakdown [“moderate lesion”]).
The metabolomics analysis yielded a total of 503 biochemical metabolites of known identity, which were carried forward to statistical analyses in tandem and individually (Table 1). Ten ECC traits showed significant correlations with the metabolome (i.e., all metabolites), the strongest one being for the localized classic ECC definition (dmfs3 >0; dCor P = 8 × 10−3), followed by the quantitative caries experience on these 5 surfaces (dmfs3, dCor P = 1.9 × 10−2). Of note, all untreated disease traits also showed significant correlations with the metabolome. In terms of individual metabolite associations, numerous associations were found to be differentially abundant in ECC traits at a nominal statistical significance level after adjustment for participants’ age and race/ethnicity. Most associations were positive (i.e., upregulations/enrichments; e.g., 52 versus 14 negative associations for the localized ECC case definition). The number of unrestored caries lesions (at the ICDAS ≥3 threshold) was the trait with the largest dCor value (dCor = 0.230).
Sixteen metabolites remained statistically significantly associated with ECC after FDR correction (Table 2), and 5 of those were associated with 2 traits. These identified biochemicals included microbial and host metabolites, as well as exogenous substances. As illustrated in Figure 1D, catechin and epicatechin were inversely associated with the localized classic ECC definition (dmfs3 >0) and the corresponding quantitative caries experience trait (dmfs3), while fucose and N-acetylneuraminate were positively associated with the same traits. The magnitude of these associations remained virtually unchanged after adjustment for participants’ age and race/ethnicity (Table 2). The joint distribution of these metabolites’ log2-transformed abundances with the 2 ECC traits of interest, as well as correlations with participants’ characteristics, is presented in Appendix Figures 2 to 4. Additional significant positive associations with ECC were noted for imidazole propionate, 9,10-DiHOME, inosine, 3- (4-hydroxyphenyl) lactate (HPLA), 1-stearoyl-GPG, 12, 13-DiHOME, xanthine and sedoheptulose-7-phosphate, and inverse associations for raffinose and stachyose (Appendix Figs. 5–8). Bivariate association results for creatine and creatinine with ECC are presented in Appendix Figure 9.
Table 2.
Crude and Age- and Race/Ethnicity-Adjusted Estimates Association for the 16 Metabolites Significantly Altered with ECC Traits after False Discovery Rate Correction.
| Metabolite | Origin | ECC Trait | β (P Value), Crude | β (P Value), Age and Race/Ethnicity Adjusted | TPOT ML Feature Coefficient | TPOT ML Feature Coefficient Rank |
|---|---|---|---|---|---|---|
| Catechin | Exogenous | dmfs3 >0 | −0.652 (4.7 × 10−6) | −0.704 (4.3 × 10−6) | 0.011 | 1 |
| dmfs3 | −0.195 (5.2 × 10−5) | −0.216 (3.1 × 10−5) | ||||
| Epicatechin | Exogenous | dmfs3 >0 | −0.660 (2.9 × 10−6) | −0.629 (3.6 × 10−5) | 0.0087 | 2 |
| dmfs3 | −0.213 (7.3 × 10−6) | −0.204 (7.0 × 10−5) | ||||
| Fucose | Microbial/host/exogenous | dmfs3 >0 | 0.537 (3.0 × 10−6) | 0.526 (2.3 × 10−5) | 0.0048 | 9 |
| dmfs3 | 0.157 (5.6 × 10−5) | 0.150 (3.6 × 10−4) | ||||
| N-acetylneuraminate | Host | dmfs3 >0 | 0.799 (6.8 × 10−6) | 0.586 (1.8 × 10−3) | 0.0043 | 14 |
| dmfs3 | 0.215 (3.5 × 10−4) | 0.132 (3.7 × 10−2) | ||||
| Creatine | Host/exogenous | dmfs3 >0 | 0.448 (8.4 × 10−4) | 0.359 (1.2 × 10−2) | 0.0033 | 21 |
| dmfs >0 | 0.544 (3.1 × 10−5) | 0.540 (7.5 × 10−5) | ||||
| Creatinine | Host | dmfs >0 | 0.586 (2.0 × 10−4) | 0.625 (1.3 × 10−4) | ||
| Imidazole propionate | Microbial | dmfs3 >0 | 0.682 (5.3 × 10−4) | 0.589 (5.5 × 10−3) | 0.005 | 8 |
| 9,10-DiHOME | Exogenous/microbial | dmfs3 >0 | 0.583 (2.2 × 10−4) | 0.675 (7.4 × 10−5) | 0.0047 | 11 |
| Inosine | Microbial/host | dmfs3 >0 | 0.565 (6.0 × 10−5) | 0.449 (2.8 × 10−3) | 0.0032 | 22 |
| 3-(4-Hydroxyphenyl) lactate (HPLA) | Host | dmfs3 >0 | 0.449 (4.2 × 10−4) | 0.332 (1.4 × 10−2) | 0.0024 | 36 |
| 1-Stearoyl-GPG | Host | dmfs3 >0 | 0.550 (9.2 × 10−4) | 0.312 (6.8 × 10−2) | 0.0019 | 47 |
| 12,13-DiHOME | Exogenous/microbial | dmfs3 >0 | 0.503 (7.3 × 10−4) | 0.567 (4.0 × 10−4) | 0.001 | 81 |
| Xanthine | Host/exogenous/ microbial | dmfs3 >0 | 0.725 (1.0 × 10−3) | 0.492 (3.5 × 10−2) | −0.0007 | 204 |
| Raffinose | Exogenous | dmfs3 >0 | −0.758 (1.0 × 10−3) | −0.739 (3.0 × 10−3) | −0.0012 | 256 |
| Stachyose | Exogenous | dmfs3 >0 | −0.722 (6.6 × 10−4) | −0.674 (3.1 × 10−3) | −0.0027 | 402 |
| Sedoheptulose-7-phosphate | Microbial/host | dmfs3 >0 | 0.475 (6.5 × 10−4) | 0.301 (3.7 × 10−2) | −0.0033 | 452 |
ML-derived feature importance coefficients and ranks are presented for the 15 metabolites that showed significant associations with the binary localized ECC trait (i.e., dmfs3 >0 defined at the ICDAS 1 threshold).
dmfs, the sum of decayed, missing, and restored (i.e., “filled”) primary tooth surfaces due to caries; ECC, early childhood caries; ICDAS, International Caries Detection and Classification System; ML, machine learning.
The TPOT approach outperformed all competitive automated ML strategies; the best-performing model for the localized classic ECC case status classification consisted of a logistic regression classifier and 2 feature transformers (i.e., robust scaler and stacking estimator with the k-neighbor classifier) and had 66% balanced accuracy. The final model had modest predictive performance (area under the receiver operating characteristic [ROC] curve = 0.75); nevertheless, it demonstrated the discriminant potential of several of the identified significantly altered metabolites (Table 2). Catechin had the highest ECC classification importance (i.e., mean decrease in model accuracy = 1.1%) and epicatechin was second, whereas fucose, imidazole propionate, 9,10-DiHOME, and N-acetylneuraminate were among the top 15 metabolites in terms of ECC classification importance (Fig. 2).
Figure 2.
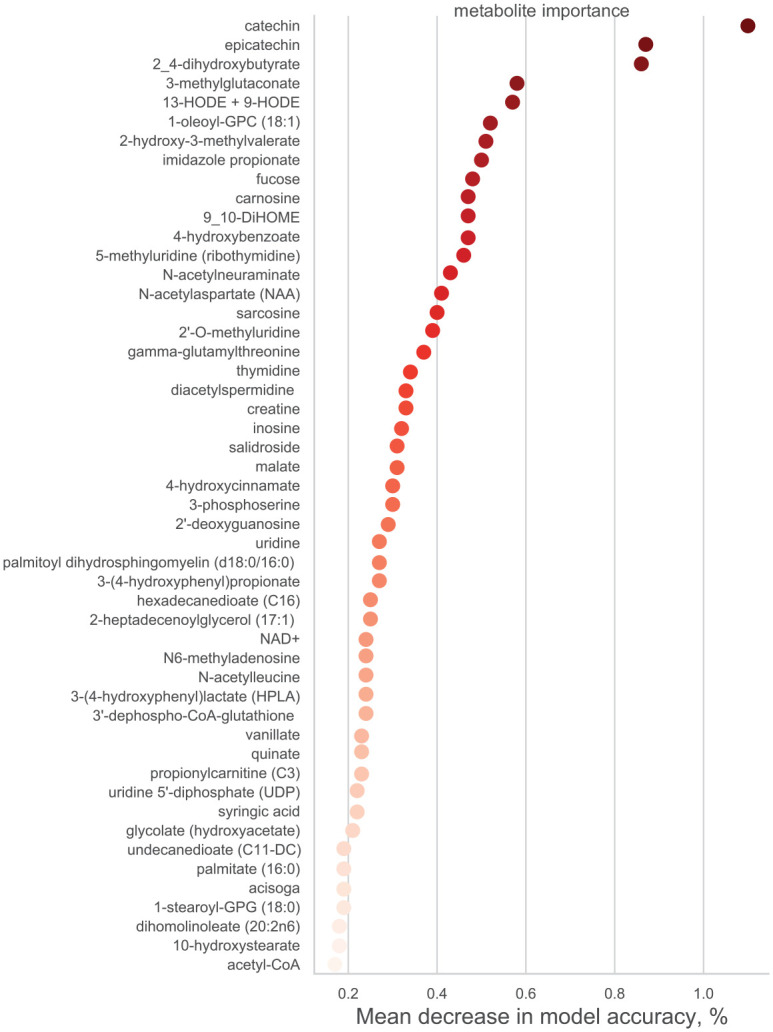
Feature importance plot for the top 50 metabolites in the best-fitting tree-based pipeline optimization tool (TPOT) AutoML model. The metabolites are presented in order of descending “feature importance” in the AutoML model. The result for the top metabolite, catechin, can be interpreted as 1.1% relative classification performance decrease if catechin values are permuted in the TPOT prediction model.
Discussion
We carried out a comprehensive untargeted metabolomics characterization of the supragingival biofilm in a sizable sample of 289 preschool-age children and identified several altered biochemicals associated with ECC. Our results offer evidence of an overall association between the supragingival biofilm biochemical composition and ECC and highlight the roles of several endogenous and exogenous metabolites. These metabolite associations remained robust to adjustments for participants’ demographic characteristics, and most emerged as important features in an ECC ML classification model. While these findings will need to be replicated in independent samples and validated mechanistically, they provide substantial new information on the supragingival biofilm metabolome of young children and its association with ECC.
We explored global metabolome associations with 18 ECC traits, a number that may initially appear high or unwarranted; however, this is an important and necessary step in our understanding of ECC metabolomics, as clinical trait definitions can and do vary considerably. We interrogated 2 different levels of caries severity (i.e., including or excluding noncavitated caries lesions) and a third, untreated disease category, all of which are clinically important. We considered disease prevalence and severity on the entire dentition, as well as on the specific teeth and specific surfaces (i.e., “localized disease”) where plaque was harvested from. We posited that the biofilm metabolome may be most informative for surfaces and teeth from which it was collected, rather than the entire dentition—this notion was verified by the strongest correlations being found for the 2 localized disease traits. All untreated disease traits showed significant correlations with the metabolome, although smaller numbers of significantly altered individual metabolites and only 2 metabolites remained significantly associated with the person-level ECC definition defined at the ICDAS ≥3 threshold after correction for multiple testing.
Most identified metabolite associations were positive (i.e., ECC prevalence and severity were associated with higher relative metabolite abundance), a finding consistent with a biochemically active microbial community in the context of dental caries. Nevertheless, 2 of the strongest observed associations, for catechin and epicatechin, were negative. Both catechin and epicatechin are known dietary flavonoids (i.e., exogenous substances), a class of polyphenolic plant secondary metabolites with several biological properties. They are commonly found in food sources such as blackcurrants, cranberries, cocoa powder, chocolate, and green and white tea and have shown anticaries properties, including inhibition of cariogenic bacteria adhesion, acid production, and biofilm formation (Jeon et al. 2011; Varoni et al. 2012; Li et al. 2019). A recent review by Hengge (2019) offers an insightful summary of the postulated mechanisms of a green tea polyphenol catechin in antagonizing bacterial biofilms, which may involve alterations of the pellicle (Rehage et al. 2017).
Fucose is present in host-derived glycoproteins, and it can be produced by bacteria and acquired from the diet (Becker and Lowe 2003). Its functional role in the oral cavity is currently unclear; however, it can be used by certain fucosidase-expressing streptococci and appears to mediate bacterial binding, including early streptococcal colonizers via antigen I/II adhesins (Cross and Ruhl 2018; You et al. 2019). Interestingly, free fucose has been shown to inhibit saliva-mediated aggregation and clearance of Streptococcus mutans (Demuth et al. 1990), potentially enhancing its adherence and accumulation on tooth surfaces. N-acetylneuraminate is the conjugate base of N-acetylneuraminic acid, the most abundant sialic acid in humans. Sialic acids are important in terms of host immunity regulation and biological functions of health and disease-associated bacteria (Severi et al. 2007).
There is unavoidable and unobserved complexity in what is sampled and measured in observational supragingival biofilm metabolomics studies like the present one. For example, the origins of the metabolome: Schulz et al. (2020) recently conducted targeted metabolomics analyses of initially formed (10 min) pellicle in young children and found that the identified metabolites were not significantly different from what was quantified in saliva. Importantly, the most abundant metabolites, including acetic acid, propionic acid, glycine, serine, galactose and mannose, lactose, glucose, palmitic acid, and stearic acid, were identified in the virtual absence of bacterial colonization. Examinations of the temporal development and contributions to the biofilm metabolome are logical areas for future study.
The performance of the best-fitting automated ML model showed that the metabolome alone did not contain sufficient information to accurately classify (i.e., distinguish between) ECC cases and noncases in our study. This is not surprising given the amount of heterogeneity encompassed within the current ECC taxonomy in terms of severity and intraoral distribution of caries lesions (Divaris 2016). However, the ML model independently prioritized several metabolites that emerged as significant in conventional statistical analyses of ECC among the top “predictive” ones, including catechin, epicatechin, imidazole propionate, fucose, 9,10-DiHOME, and inosine. Others (i.e., xanthine, raffinose, stachyose, and sedoheptulose-7-phosphate) were noncontributory to ECC classification. It is conceivable that in the future, information on a small number of features (versus full information on all omics), whether metabolites, microbial taxa, behaviors, or screening questions, may be efficiently used by automated ML pipelines to create useful clinical decision-making adjuncts for ECC classification or risk estimation.
This cross-sectional study is limited in its ability to make causal or true predictive inferences because it examined prevalent ECC. This is particularly true for untreated disease, wherein the clinical manifestations of ECC (i.e., cavitation) alter the microbial niche and biofilm function. The pooling of plaque samples from 5 specific tooth surfaces provided uniformity across the entire study sample but limited our ability to link the metabolite associations with site-specific ECC manifestations. Future studies can and must expand upon our findings by investigating site-specific variation and longitudinal changes in biofilm metabolic activity. Finally, it was not practically possible to collect fasting plaque (i.e., plaque unexposed to sugar for ≥12 h) or record the breakfast sugar content in this community-based, observational study among children aged 3 to 5 y. Plaque samples were collected prior to or at least 30 min after breakfast and—importantly with regards to study validity—all participants were exposed to largely similar conditions in the state’s public preschool system, while identical timing and plaque collection conditions applied to both ECC cases and controls.
In sum, our results point to several novel metabolite associations with ECC, with plausible biological roles that need to be mechanistically validated. Certainly, these metabolites do not operate in a vacuum, and future studies should investigate their associations and potential interactions with the microbial community structure and activity (i.e., metagenomics, metatranscriptomics). Crucially, the measured supragingival biofilm biochemicals contained both host and exogenous substances, a demonstration of its complexity and unexplored potential beyond microbiota-derived metabolites. Several novel metabolite associations could be combined and serve as ECC biomarkers in the future; however, the information currently contained in the metabolome alone appears insufficient to accurately differentiate ECC cases from noncases. Our ability to accurately classify or predict ECC outcomes will likely improve with the addition and joint consideration of additional levels of microbial omics, host genomics, behavioral and environmental factors, and a precise ECC taxonomy in the context of longitudinal studies.
Author Contributions
L.H. Heimisdottir, contributed to design and data interpretation, drafted and critically revised the manuscript; B.M. Lin, A. Orlenko, H. Koo, contributed to data analysis and interpretation, drafted and critically revised the manuscript; H. Cho, contributed to data analysis, critically revised the manuscript; A.A. Ribeiro, D. Shungin, contributed to data interpretation, critically revised the manuscript; A. Simon-Soro, J. Roach, H.D. Spangler, J.H. Moore, contributed to data analysis and interpretation, critically revised the manuscript; J. Ginnis, M.A. Simancas-Pallares, contributed to conception and data acquisition, critically revised the manuscript; A.G. Ferreira Zandoná, contributed to conception, design, and data interpretation, critically revised the manuscript; J.T. Wright, contributed to data interpretation, drafted and critically revised the manuscript; P. Ramamoorthy, contributed to design, data acquisition, analysis, and interpretation, critically revised the manuscript; D. Wu, contributed to design, data analysis, and interpretation, critically revised the manuscript; K. Divaris, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, sj-pdf-1-jdr-10.1177_0022034520982963 for Metabolomics Insights in Early Childhood Caries by L.H. Heimisdottir, B.M. Lin, H. Cho, A. Orlenko, A.A. Ribeiro, A. Simon-Soro, J. Roach, D. Shungin, J. Ginnis, M.A. Simancas-Pallares, H.D. Spangler, A.G. Ferreira Zandoná, J.T. Wright, P. Ramamoorthy, J.H. Moore, H. Koo, D. Wu and K. Divaris in Journal of Dental Research
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
sj-xlsx-2-jdr-10.1177_0022034520982963 for Metabolomics Insights in Early Childhood Caries by L.H. Heimisdottir, B.M. Lin, H. Cho, A. Orlenko, A.A. Ribeiro, A. Simon-Soro, J. Roach, D. Shungin, J. Ginnis, M.A. Simancas-Pallares, H.D. Spangler, A.G. Ferreira Zandoná, J.T. Wright, P. Ramamoorthy, J.H. Moore, H. Koo, D. Wu and K. Divaris in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study was supported by grants from the National Institutes of Health/National Institute of Dental and Craniofacial Research (NIH/NIDCR) U01DE025046, R03DE028983, R01DE025220.
ORCID iDs: L.H. Heimisdottir  https://orcid.org/0000-0002-7630-8883
https://orcid.org/0000-0002-7630-8883
K. Divaris  https://orcid.org/0000-0003-1290-7251
https://orcid.org/0000-0003-1290-7251
References
- Becker DJ, Lowe JB. 2003. Fucose: biosynthesis and biological function in mammals. Glycobiology. 13(7):41R–53R. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 57(1):289–300. [Google Scholar]
- Bowen WH, Burne RA, Wu H, Koo H. 2018. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26(3):229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. 2001. Random forests. Mach Learn. 45(1):5–32. [Google Scholar]
- Casamassimo PS, Thikkurissy S, Edelstein BL, Maiorini E. 2009. Beyond the dmft: the human and economic cost of early childhood caries. J Am Dent Assoc. 140(6):650–657. [DOI] [PubMed] [Google Scholar]
- Cross BW, Ruhl S. 2018. Glycan recognition at the saliva - oral microbiome interface. Cell Immunol. 333:19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth DR, Lammey MS, Huck M, Lally ET, Malamud D. 1990. Comparison of Streptococcus mutans and Streptococcus sanguis receptors for human salivary agglutinin. Microb Pathog. 9(3):199–211. [DOI] [PubMed] [Google Scholar]
- Divaris K, Joshi A. 2020. The building blocks of precision oral health in early childhood: the ZOE 2.0 study. J Public Health Dent. 80(Suppl 1):S31–S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divaris K, Slade GD, Ferreira Zandona AG, Preisser JS, Ginnis J, Simancas-Pallares MA, Agler CS, Shrestha P, Karhade DS, Ribeiro AA, et al. 2020. Cohort profile: ZOE 2.0—a community-based genetic epidemiologic study of early childhood oral health. Int J Environ Res Public Health. 17(21):8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divaris K, Shungin D, Rodríguez-Cortés A, Basta PV, Roach J, Cho H, Wu D, Ferreira Zandoná AG, Ginnis J, Ramamoorthy S, et al. 2019. The supragingival biofilm in early childhood caries: clinical and laboratory protocols and bioinformatics pipelines supporting metagenomics, metatranscriptomics, and metabolomics studies of the oral microbiome. Methods Mol Biol. 1922:525–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divaris K. 2016. Predicting dental caries outcomes in children: a “risky” concept. J Dent Res. 95(3):248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. 2009. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 81(16):6656–6667. [DOI] [PubMed] [Google Scholar]
- Evans AM, Bridgewater BR, Liu Q, Mitchell MW, Robinson RJ, Dai H, Stewart SJ, DeHaven CD, Miller LA. 2014. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics. 4:2. [Google Scholar]
- Ginnis J, Ferreira Zandoná AG, Slade GD, Cantrell J, Antonio ME, Pahel BT, Meyer BD, Shrestha P, Simancas-Pallares MA, Joshi AR, et al. 2019. Measurement of early childhood oral health for research purposes: dental caries experience and developmental defects of the enamel in the primary dentition. Methods Mol Biol. 1922:511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis E, Parsaei Y, Klein MI, Koo H. 2017. Advances in the microbial etiology and pathogenesis of early childhood caries. Mol Oral Microbiol. 32(1):24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug K, Cochrane K, Nainala VC, Williams M, Chang J, Jayaseelan KV, O’Donovan C. 2020. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 48(D1):D440–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. 2019. Targeting bacterial biofilms by the green tea polyphenol EGCG. Molecules. 24(13):2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley E, Barrett MPJ, Kinirons M, Whelton H, Ryan CA, Stanton C, Harris HMB, O’Toole PW. 2019. Comparison of the salivary and dentinal microbiome of children with severe-early childhood caries to the salivary microbiome of caries-free children. BMC Oral Health. 19(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JG, Rosalen PL, Falsetta ML, Koo H. 2011. Natural products in caries research: current (limited) knowledge, challenges and future perspective. Caries Res. 45(3):243–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke T, Kneist S, de Soet JJ, Kuhlisch E, Mauersberger S, Forster A, Klimm W. 2009. Acid production by oral strains of Candida albicans and lactobacilli. Caries Res. 43(2):83–91. [DOI] [PubMed] [Google Scholar]
- Li Y, Jiang X, Hao J, Zhang Y, Huang R. 2019. Tea polyphenols: application in the control of oral microorganism infectious diseases. Arch Oral Biol. 102:74–82. [DOI] [PubMed] [Google Scholar]
- Mira A. 2018. Oral Microbiome studies: potential diagnostic and therapeutic implications. Adv Dent Res. 29(1):71–77. [DOI] [PubMed] [Google Scholar]
- Nascimento MM, Zaura E, Mira A, Takahashi N, Ten Cate JM. 2017. Second era of OMICS in caries research: moving past the phase of disillusionment. J Dent Res. 96(7):733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyvad B, Crielaard W, Mira A, Takahashi N, Beighton D. 2013. Dental caries from a molecular microbiological perspective. Caries Res. 47(2): 89–102. [DOI] [PubMed] [Google Scholar]
- Olson RS, Moore JH. 2019. TPOT: a tree-based pipeline optimization tool for automating machine learning. In: Hutter F, Kotthoff L, Vanschoren J, editors. Automated machine learning: methods, systems, challenges. Cham (Switzerland): Springer. p. 151–160. [Google Scholar]
- Orlenko A, Kofink D, Lyytikäinen LP, Nikus K, Mishra P, Kuukasjärvi P, Karhunen PJ, Kähönen M, Laurikka JO, Lehtimäki T, et al. 2020. Model selection for metabolomics: predicting diagnosis of coronary artery disease using automated machine learning. Bioinformatics. 36(6):1772–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts NB, Baez RJ, Diaz-Guillory C, Donly KJ, Alberto Feldens C, McGrath C, Phantumvanit P, Seow WK, Sharkov N, Songpaisan Y, et al. 2019. Early childhood caries: IAPD Bangkok Declaration. J Dent Child (Chic). 86(2):72. [PubMed] [Google Scholar]
- Pitts NB, Zero DT, Marsh PD, Ekstrand K, Weintraub JA, Ramos-Gomez F, Tagami J, Twetman S, Tsakos G, Ismail A. 2017. Dental caries. Nat Rev Dis Primers. 3:17030. [DOI] [PubMed] [Google Scholar]
- Rehage M, Delius J, Hofmann T, Hannig M. 2017. Oral astringent stimuli alter the enamel pellicle’s ultrastructure as revealed by electron microscopy.J Dent. 63:21–29. [DOI] [PubMed] [Google Scholar]
- Rosier BT, Marsh PD, Mira A. 2018. Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J Dent Res. 97(4):371–380. [DOI] [PubMed] [Google Scholar]
- Schulz A, Lang R, Behr J, Hertel S, Reich M, Kümmerer K, Hannig M, Hannig C, Hofmann T. 2020. Targeted metabolomics of pellicle and saliva in children with different caries activity. Sci Rep. 10(1):697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severi E, Hood DW, Thomas GH. 2007. Sialic acid utilization by bacterial pathogens. Microbiology. 153(9):2817–2822. [DOI] [PubMed] [Google Scholar]
- Székely GJ, Rizzo ML. 2009. Brownian distance covariance. Ann Appl Stat. 3(4):1236–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoni EM, Lodi G, Sardella A, Carrassi A, Iriti M. 2012. Plant polyphenols and oral health: old phytochemicals for new fields. Curr Med Chem. 19(11):1706–1720. [DOI] [PubMed] [Google Scholar]
- Wei R, Wang J, Su M, Jia E, Chen S, Chen T, Ni Y. 2018. Missing value imputation approach for mass spectrometry-based metabolomics data. Sci Rep. 8(1):663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Grier A, Faustoferri RC, Alzoubi S, Gill AL, Feng C, Liu Y, Quivey RG, Kopycka-Kedzierawski DT, Koo H, et al. 2018. Association between oral candida and bacteriome in children with severe ECC. J Dent Res. 97(13):1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim S, Yildiz E, Kubar A. 2010. TaqMan real-time quantification of Epstein-Barr virus in severe early childhood caries. Eur J Dent. 4(1):28–33. [PMC free article] [PubMed] [Google Scholar]
- You J, Lin S, Jiang T. 2019. Origins and evolution of the α-L-fucosidases: from bacteria to metazoans. Front Microbiol. 10:1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP;, STROBE Initiative. 2007. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet. 370(9596):1453–1457. [DOI] [PubMed] [Google Scholar]
- Zandona F, Soini HA, Novotny MV, Santiago E, Eckert GJ, Preisser JS, Benecha HK, Arthur RA, Zero DT. 2015. A potential biofilm metabolite signature for caries activity—a pilot clinical study. Metabolomics. 5(1):140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jdr-10.1177_0022034520982963 for Metabolomics Insights in Early Childhood Caries by L.H. Heimisdottir, B.M. Lin, H. Cho, A. Orlenko, A.A. Ribeiro, A. Simon-Soro, J. Roach, D. Shungin, J. Ginnis, M.A. Simancas-Pallares, H.D. Spangler, A.G. Ferreira Zandoná, J.T. Wright, P. Ramamoorthy, J.H. Moore, H. Koo, D. Wu and K. Divaris in Journal of Dental Research
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
sj-xlsx-2-jdr-10.1177_0022034520982963 for Metabolomics Insights in Early Childhood Caries by L.H. Heimisdottir, B.M. Lin, H. Cho, A. Orlenko, A.A. Ribeiro, A. Simon-Soro, J. Roach, D. Shungin, J. Ginnis, M.A. Simancas-Pallares, H.D. Spangler, A.G. Ferreira Zandoná, J.T. Wright, P. Ramamoorthy, J.H. Moore, H. Koo, D. Wu and K. Divaris in Journal of Dental Research