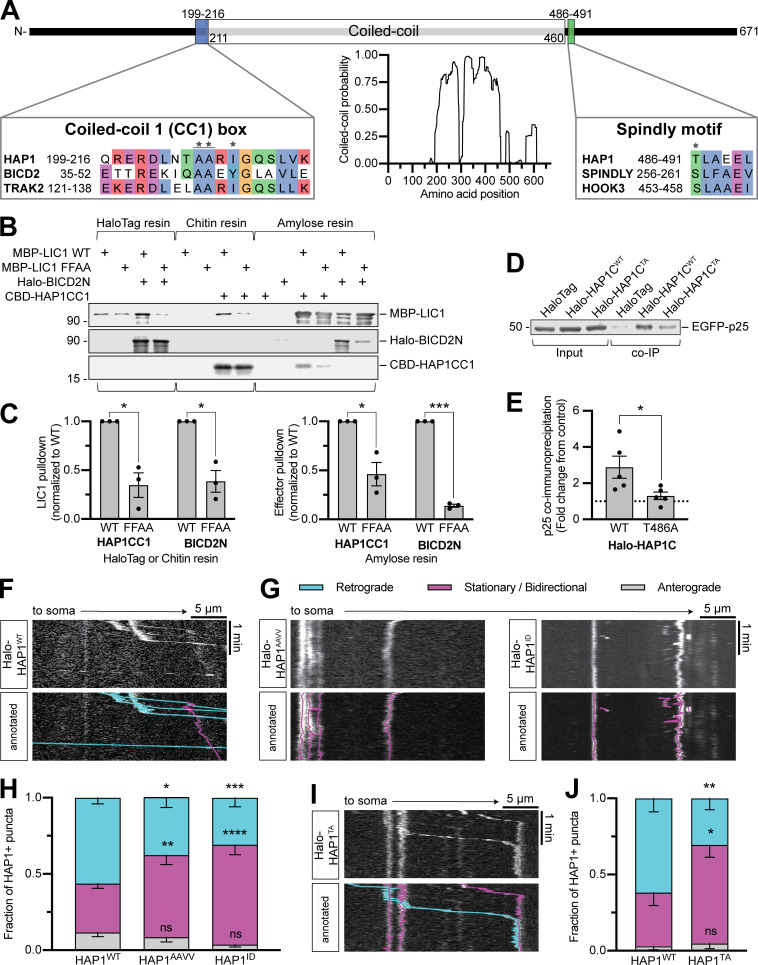
Figure 3.
HAP1 binds dynein–dynactin via activating adaptor motifs. (A) Schematic illustrating the domain architecture of HAP1. Coiled-coil probability was calculated using predictions from four different programs. Residues in the sequence alignments are colored using the clustal color scheme where conserved. Stars indicate point mutants. (B and C) Immunoblot and quantification of purified HAP1CC1 (aa 168–261) or BICD2N (aa 1–572) pulldown by dynein LIC1WT or mutant LIC1FFAA and vice versa. Results were normalized to the LIC1WT condition; n = 3 pulldowns from n = 2 independent purifications; paired two-tailed t test; LIC1 pulldown (HAP1CC1 WT vs. FFAA, P = 0.0353; BICD2N WT vs. FFAA, P = 0.0051); effector pulldown (HAP1CC1 WT vs. FFAA, P = 0.0109; BICD2N WT vs. FFAA, P < 0.0001). (D and E) Example immunoblot and quantification of EGFP-p25 coimmunoprecipitation by HAP1CWT (aa 470–671) or mutant HAP1CTA (coexpressed in COS-7 cells). Results were normalized to p25 pulled down by HaloTag only (negative control; dotted black line); n = 5 repeats; paired t test (two-tailed; P = 0.0450). (F–J) Example kymographs and quantification of motile behavior of punctate HAP1WT (F), HAP1 CC1 box (G and H), and HAP1 Spindly (I and J) mutants. n = 11–16 neurons; two-way ANOVA with Bonferroni’s multiple comparisons test; AAVV (retrograde, P = 0.0333; stationary/bidirectional [Stat/Bidir], P = 0.0099); ID (retrograde, P = 0.0009; Stat/Bidir, P < 0.0001); TA (retrograde, P = 0.0063; Stat/Bidir, P = 0.0109). Bars throughout show mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 compared to WT.