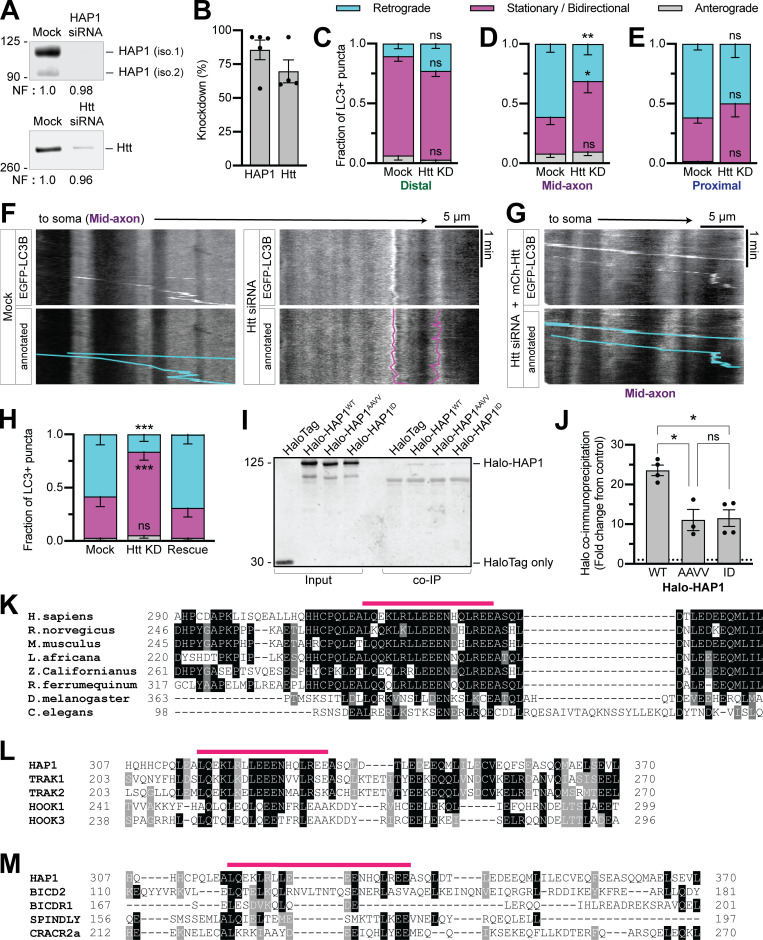
Figure S2.
Htt regulates autophagosomal motility in the mid-axon and HAP1 contains conserved dynein–dynactin binding sites. (A and B) Immunoblotting and quantification of PC12 cell lysates show KD efficiency of HAP1 and Htt siRNAs. Normalization factor (NF) determined using Revert Total Protein Stain. n = 4–5 repeats. (C–E) Quantification of LC3+ puncta motile behavior in the distal, mid-, and proximal axonal regions. n = 6–21 neurons. Two-way ANOVA with Sidak’s multiple comparisons test (mid retrograde mock vs. Htt KD, P = 0.0076; mid stationary/bidirectional [Stat/Bidir] mock vs. Htt KD, P = 0.0132). (F) Example kymographs from the mid-axon of a mock-transfected (control) neuron and a neuron transfected with Htt siRNA. (G and H) Example kymograph and quantification from the mid-axon of a neuron transfected with Htt siRNA and siRNA-resistant mCh-Htt. Two-way ANOVA with Tukey’s multiple comparisons test (retrograde mock vs. Htt KD, P = 0.0005 = 4; Stat/Bidir mock vs. Htt KD, P = 0.0009). Symbols indicate comparison to mock. (I and J) Immunoblot and quantification of purified LIC1 pulldown by HAP1 constructs (COS-7 lysate). n = 3–4 repeats; results normalized to HaloTag only pulldown (negative control; dotted black line); mixed-effects model (fixed effect, P = 0.0233) with Tukey’s multiple comparisons test (WT vs. AAVV, P = 0.0234; WT vs. ID, P = 0.0373; AAVV vs. ID, P = 0.9936). (K) Sequence alignment of HAP1 across species. (L and M) Alignment of human HAP1 with known and putative dynein-activating adaptors (human canonical sequences). Alignments run with T-Coffee (version 11.00.d625267) default settings. Prepared for visualization by BoxShade (ExPAy, Swiss Institute of Bioinformatics) with RTF_new output. Pink lines indicate Glued motif. Black boxes indicate aa identical in ≥60% of sequences. Gray boxes indicate aa similar in ≥ 60% of sequences. Bars throughout show mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.