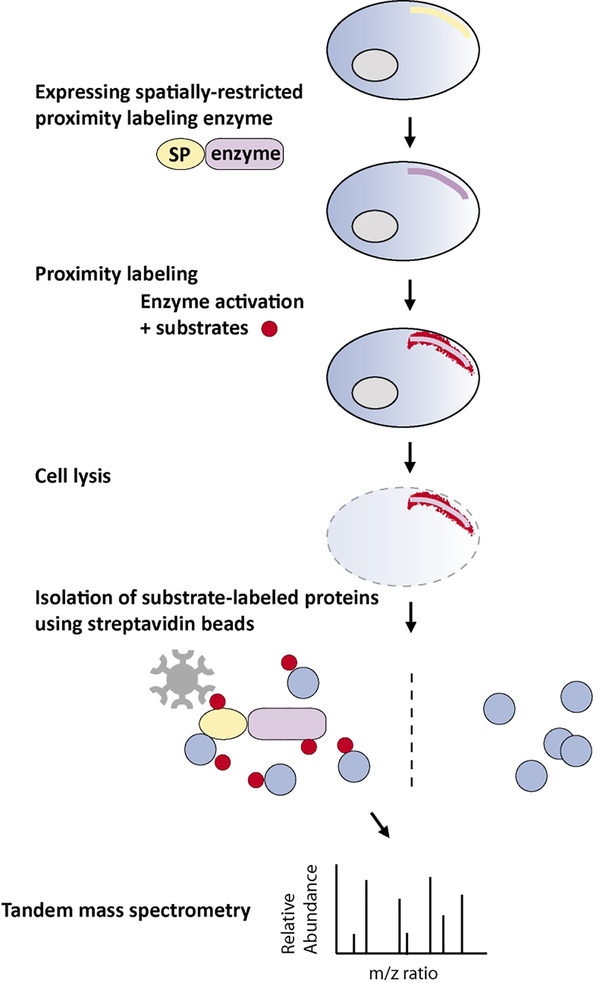
Figure 1. Proximity labeling for proteomic profiling.
To achieve regional protein labeling, the enzymes are usually fused with a targeting signal peptide or a spatially-restricted protein (SP). The enzymes can also be fused with any protein of interest for protein interactome studies. After performing proximity labeling in living cells, the cells are lysed and the tagged endogenous proteins are isolated using steptavidin beads. Small peptides of enriched proteins are generated by trypsin digestion and subsequently ionized for tandem mass spectrometry (MS/MS) analysis. The mass-to-charge (m/z) ratio of each peptide and their fragment ions is then used to identify peptide sequence through computational comparison against established databases.