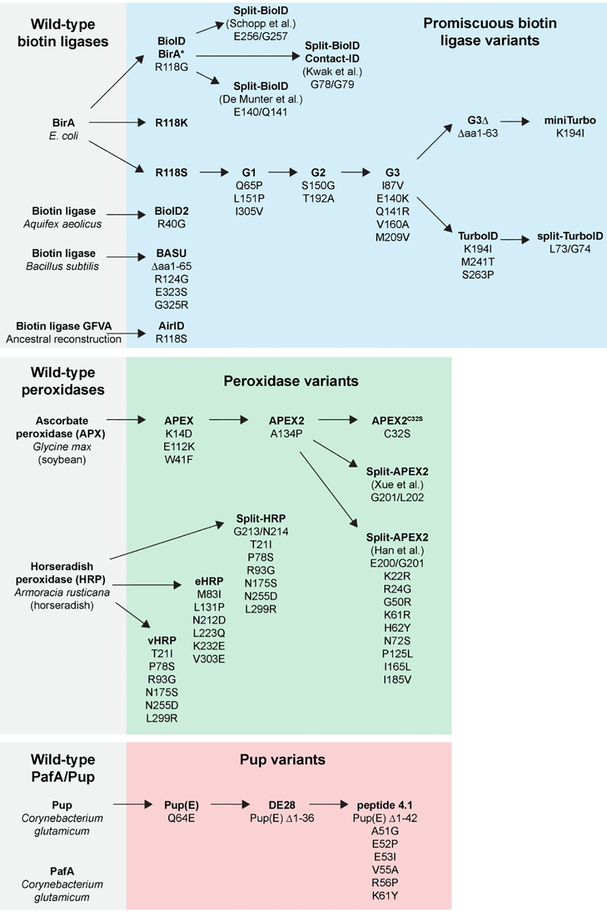
Figure 3. Directed evolution of proximity labeling components.
Proximity labeling enzymes have been modified from their wild-type counterparts by selecting for mutants with promiscuous activity. Directed evolution has been used to isolate enzymes with increased activity, increased stability, smaller molecular weight, and that are split into inactive fragments that reconstitute activity when combined. Smaller Pup substrates have also been identified.