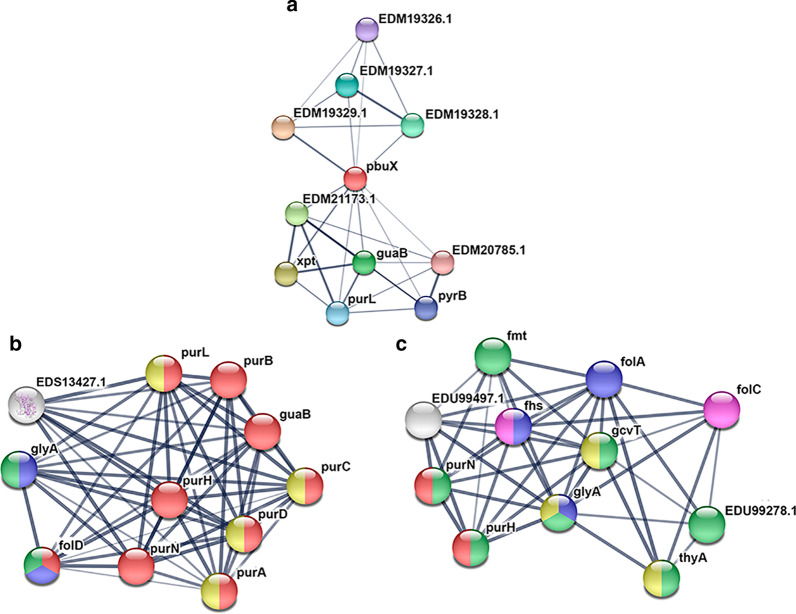
Fig. 5.
Protein–protein interaction analysis predicted by STRING. Nodes are colored based on their specific role: purine biosynthesis (red), one-carbon metabolism (blue), nucleotide-binding (yellow), ligases (pink), and amino acid biosynthesis (green). Proteins involved in multiple functions are filled with various colors. Thicker lines define the most robust associations. a Bacteroides caccae showed significant enrichment of proteins strongly involved in purine metabolism. b Bacteroides stercoris ATCC 43183, displayed functional categories included purine biosynthesis, one-carbon metabolism, nucleotide-binding, and amino-acid biosynthesis. c For Bacteroides coproccola DSM 17136 and Bacteroides stercoris ATCC 43183 we identified protein–protein interaction in enzymes involved in one-carbon metabolism, purine biosynthesis, as well as methyltransferases, transferases, and ligases