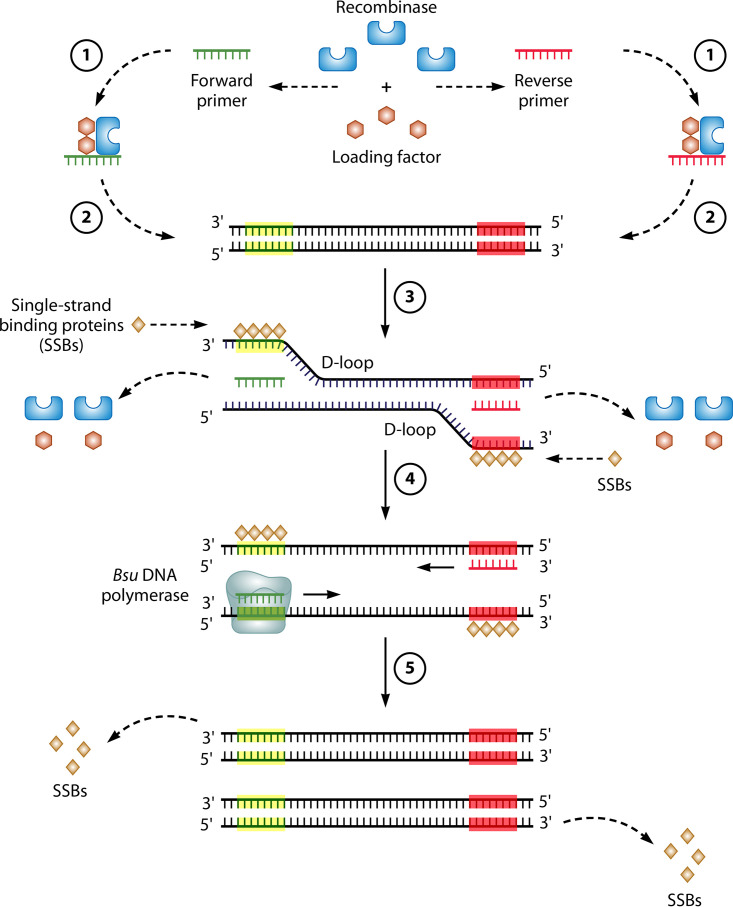
FIG 4.
Mechanism of RT-RPA. The RT-RPA reactions typically occur at between 37°C and 42°C in the following steps. (1) The reaction is initiated by the binding of a recombinase (e.g., T4 UvsX) and a loading factor (e.g., T4 UvsY) to each of the forward and reverse primers. (2) These recombinase/loading factor/oligonucleotide complexes search for homologous sequences in dsDNA, formed in the RT reaction from viral RNA (not depicted). (3) Once sequence homology is found, the recombinase complex invades the duplex DNA, forming a structure called a D-loop in an ATP-dependent reaction, where there is the unwinding of dsDNA and binding of the primer to its complementary sequence. Access to the primer-binding sequence is possible due to the stabilization of the opposite strand by SSBs (e.g., T4 gp32). Subsequently, the recombinase and loading factor disassemble and are released to initiate other rounds of target recognition. (4) Following the binding of the forward and reverse primers, these primers are extended at their 3′ ends using a strand displacement DNA polymerase (e.g., Bsu), and during the elongation process, there is a further separation of the two strands. (5) Eventually, SSBs are displaced, and the replication of both strands is complete.