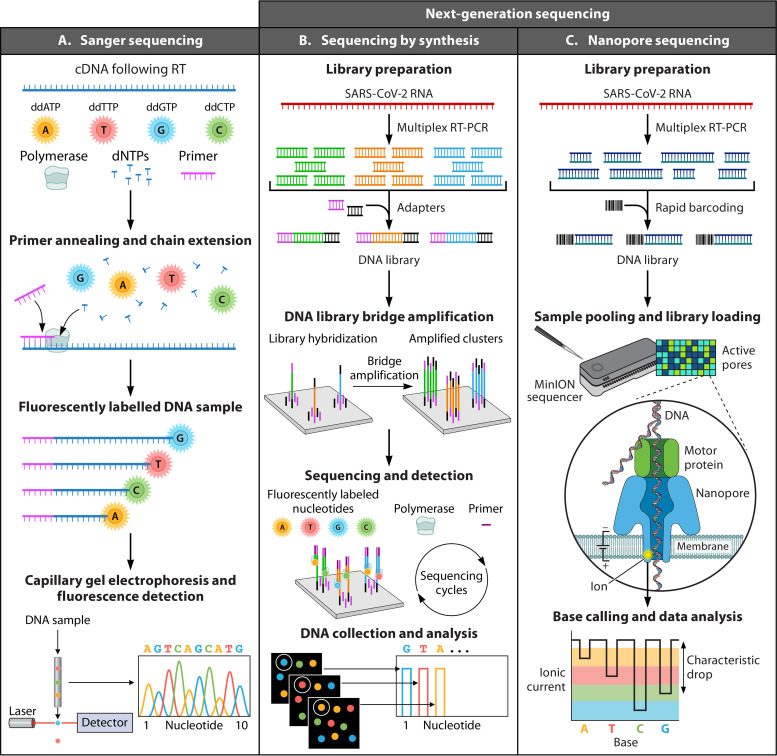
FIG 9.
Sequencing techniques for identification of SARS-CoV-2. (A) Sanger sequencing. First, SARS-CoV-2 RNA is often amplified by RT-PCR (not depicted). Sanger sequencing reactions can be undertaken to analyze either of the DNA strands, but only one strand per reaction can be assessed. The extension of the primer annealing to the template DNA occurs in the presence of DNA polymerase, buffer, cofactors, deoxynucleotide triphosphates (dNTPs), and fluorescently labeled dideoxynucleotide triphosphates (ddNTPs). The binding of the ddNTPs to the oligonucleotide strands will cease the extension, resulting in various DNA structures with different lengths. Next, the extended DNAs undergo capillary gel electrophoresis in which the shorter DNA strands move faster, resulting in the detection of the fluorescently labeled nucleotides in the order of the size of the DNA strands. Finally, as DNA fragments are resolved and nucleotide-specific fluorescence signals are captured by a detector, a chromatogram is assembled to reveal the sequence of the template. (B) Next-generation sequencing (NGS) by synthesis. First, a library of millions of DNA fragments is created from the template (or enhanced by multiplex RT-PCR for SARS-CoV-2). Adapters are bound to the two ends of each DNA fragment. The adapters consist of a universal primer-binding site and a unique sequence (i.e., barcode) that can be hybridized to a specific sequence on the support (e.g., flow cell). Following hybridization, with complementary sequences of the adapters, bridge amplification is used to amplify each DNA fragment at a defined physical position. In the sequencing and detection steps, fluorescently labeled nucleotides are bound to the forward strands in the presence of a primer and a polymerase, which results in the generation of fluorescent light that is detected by an analyzer in real time. Many other NGS technologies are also available. (C) For example, NGS by nanopore technology is presented. After creating a library of DNA fragments by multiplex RT-PCR and barcoding, the library is loaded onto a membrane containing nanopores. The nanopores are proteins that open the DNA double strand, and as each nucleotide is passed through the membrane, it causes a specific change in the ionic current that can then be translated into the nucleotide sequence of the templates.