Figure 5.
Structures of SARS-CoV-2 infectivity-enhancing antibodies bound to spike protein
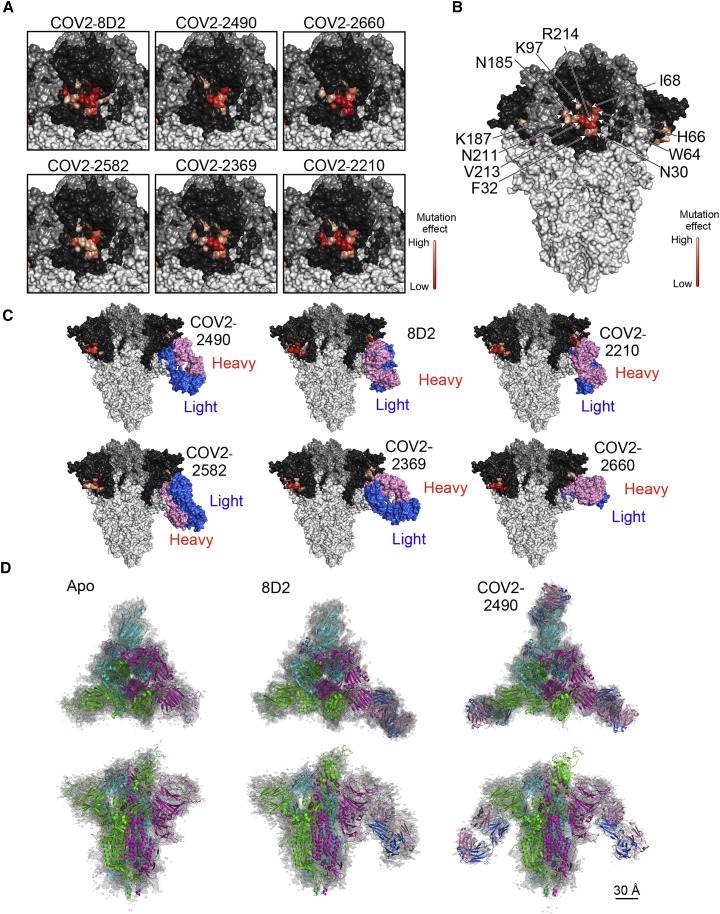
(A) Amino acid residues that affected the binding of each enhancing antibody are shown as a heatmap based on their percent reduction of the MFIs in Figure 4B, with higher reduction indicated by darker shades, as shown in bar in the figure. NTD, dark gray; RBD, medium gray; other regions, light gray.
(B) MFI reductions of the affected residues are averaged across the six antibodies and shown as a heatmap, with higher reduction indicated by darker shades, as shown in bar in the figure.
(C) Each SARS-CoV-2 infectivity-enhancing antibody was docked onto the spike protein as described in STAR Methods.
(D) A spike model built from PDB: 7KEB was found to fit the apo density best, while another model built from PDB: 7K8W fit the two other antibody-bound densities best. The spike subunit in the one RBD-“up” form is colored green. Antibodies are colored pink (heavy chain) and blue (light chain). The scale bar represents 30 Å.