Figure 3. Consensus motifs and extended features of SBS-pks and ID-pks mutational signatures.
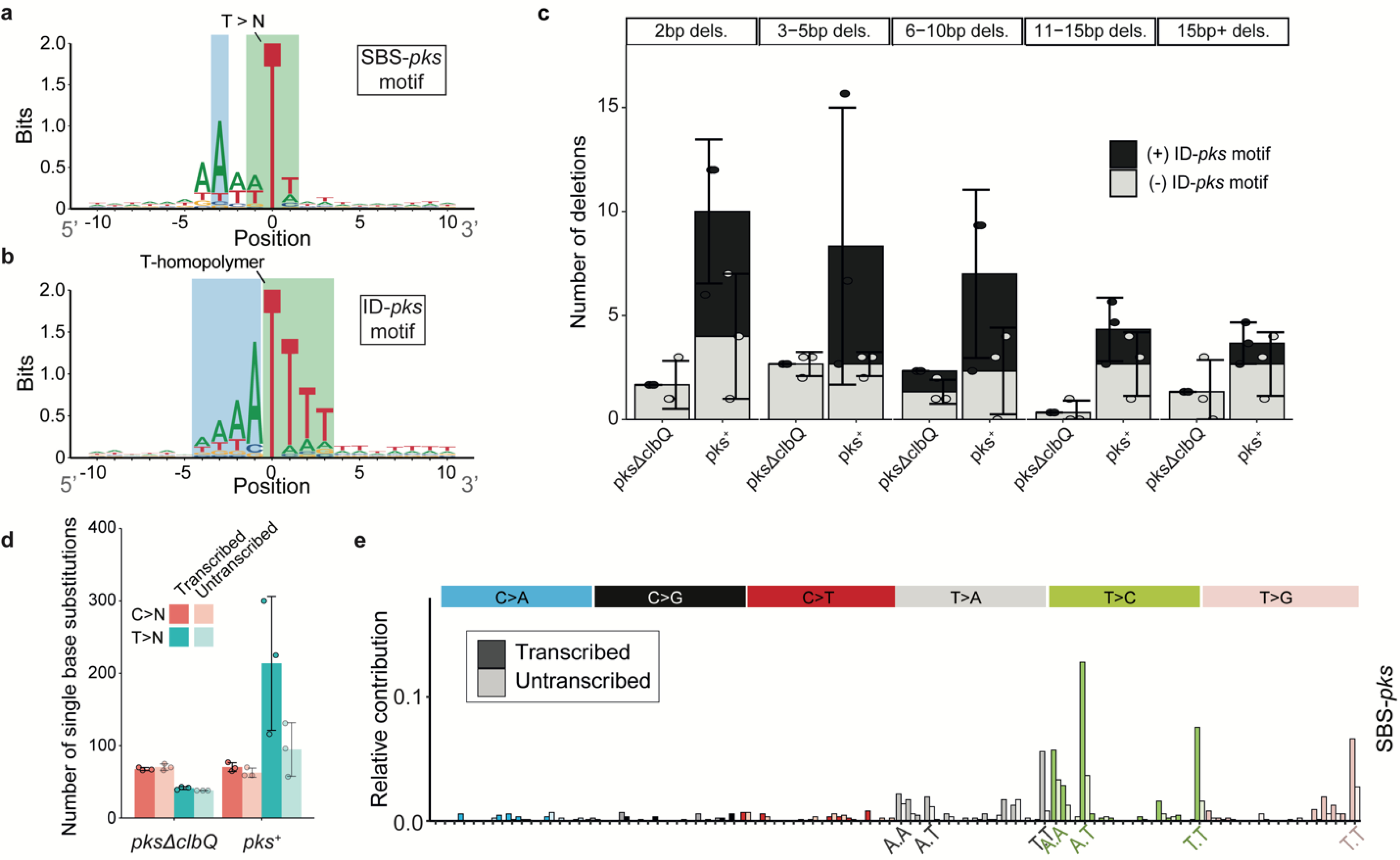
a, 2-bit representation of the extended sequence context of T>N mutations observed in organoids exposed to pks+ E. coli. Sequence directionality indicated in grey. Green: Highlighted T>N trinucleotide sequence; Blue: Highlighted A-enriched position characteristic of the SBS-pks mutations. b, 2-bit representation of the extended sequence context of single T-deletions in T-homopolymers observed in organoids exposed to pks+ E. coli. Sequence directionality indicated in grey. Green: Highlighted T-homopolymer with deleted T; Blue: Highlighted characteristic poly-A stretch. c, Mean occurrence of < 1 base pair deletions in pks+ or pksΔclbQ exposed organoids. Black bars correspond to deletions matching the ID-pks extended motifs; Grey bars correspond to deletions where no ID-pks motif is observed. Box height indicates mean number of events, error bars represent SD (n = 3 clones). d, Transcriptional strand-bias of T>N and C>N mutations occurring in organoids exposed to pks+ E. coli and pksΔclbQ E. coli. Pink: C>N; Blue: T>N; Dark color: Transcribed strand; Bright color: Untranscribed strand. Box height indicates mean number of events, error bars represent SD (n = 3 clones). e, Transcriptional strand bias of the 96-trinucleotide SBS-pks mutational signature. Color: Transcribed strand; White: Untranscribed strand.
