Extended Data Fig. 5. Signature extraction and clonal contribution of SBS-pks in CRC metastases.
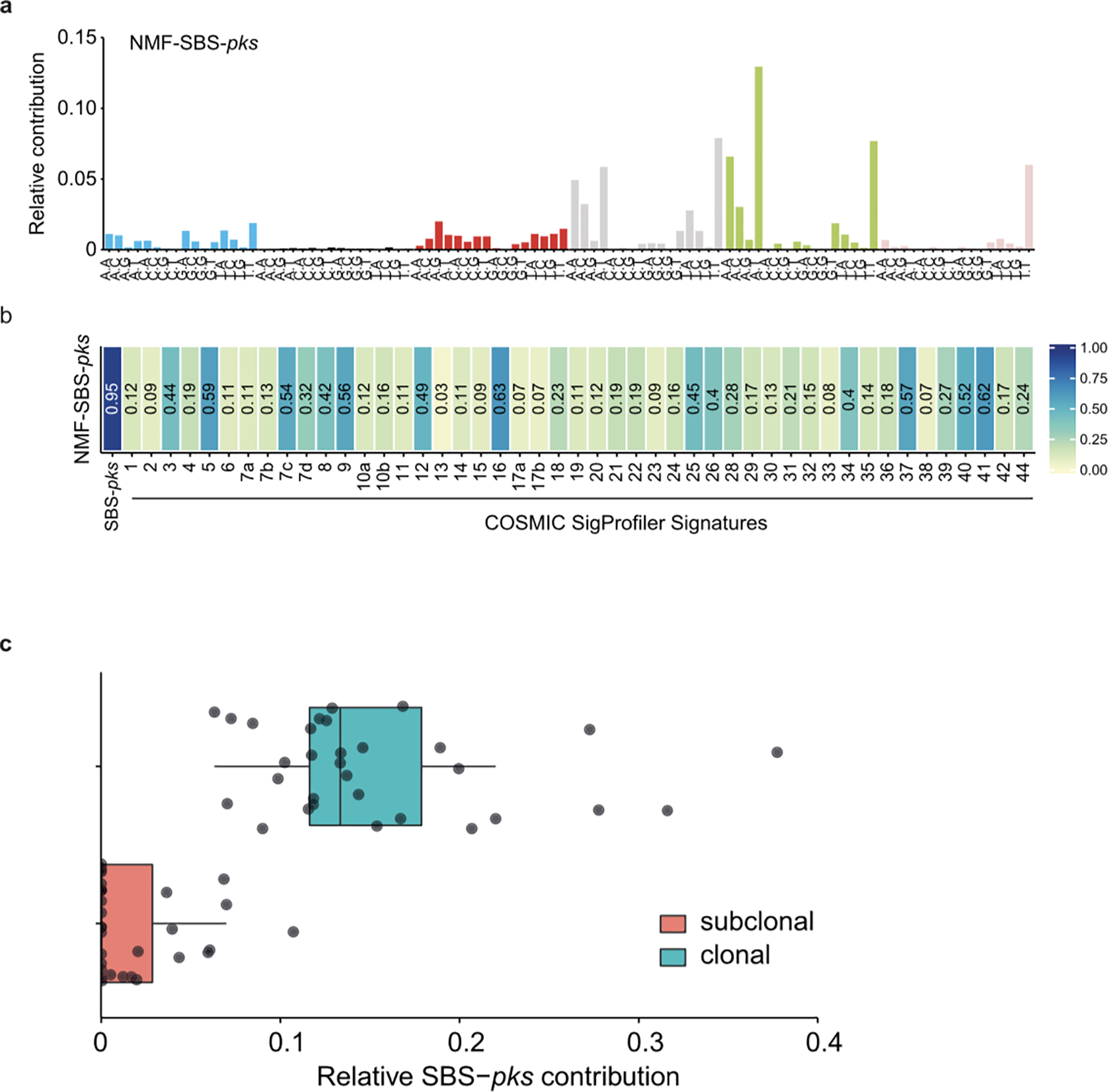
a, De-novo extracted NMF-SBS-pks signature by non-negative matrix factorization (NMF) on all 496 CRC metastases in the HMF dataset. b, Cosine similarity scores between the de-novo extracted SBS signature in (a) and COSMIC SigProfiler signatures, including our experimentally defined SBS-pks signature (left). c, Relative contribution of SBS-pks to clonal (corrected variant allele frequency > 0.4, blue bar) and subclonal fraction (corrected variant allele frequency < 0.2, red bar) of mutations in the 31 SBS/ID-pks high CRC metastases from the HMF cohort. Box indicates upper and lower quartiles with the center line indicating the mean. Box whiskers: largest value no further than 1.5 times the interquartile range extending from the box. Points indicate individual CRC metastases.
