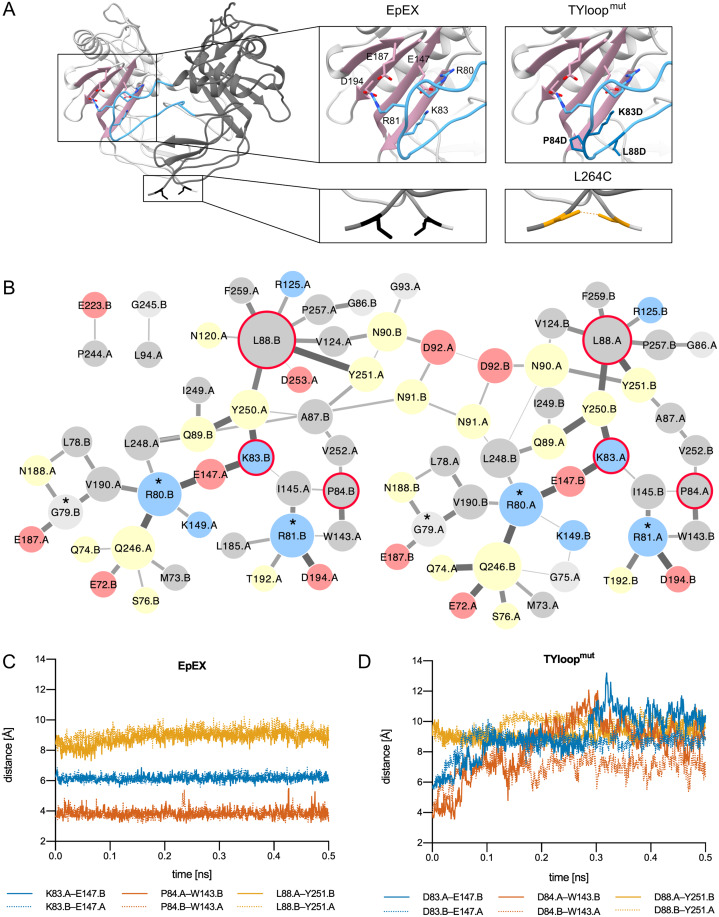
Figure 1. EpEX dimer and the designed mutants.
(A) The structure of EpEX dimer and the designed mutants. The dimer subunits are colored with different shades of grey. The β-sheet of the CD is depicted in pink and the TYloop is depicted in blue. The sidechains of key interacting residues, as inferred from the crystal structure, are shown as sticks. Residues that were mutated in TYloopmut and L264C are colored dark blue and orange, respectively. (B) Frequency of inter-subunit inter-residue contacts during EpEX MD simulation. The width of the line between two residues correlates with the frequency of contact formation—a wider line represents more frequent interaction. The size of circles corresponds to the number of interactions with other residues—a larger circle represents more interactions. Residues are colored according to their charge and polarity: positively charged blue, negatively charged red, polar yellow and non-polar grey. The residues of protease-sensitive site Gly79-Arg80-Arg81 are annotated with a star (*). Residues that were mutated in TYloopmut are encircled with a red line. (C) Changes in the key inter-subunit inter-residue distances during the first 0.5 ns of the wild type MD simulation. (D) Changes in the key inter-subunit inter-residue distances during the first 0.5 ns of the mutant TYloopmut MD simulation. For distance calculations in C and D, equivalent atoms were used for both wild type and mutated residues (Cγ for K/D83, P/D84 and L/D88); for E148 Cδ was used, and for W143 and Y251 Cζ was used.