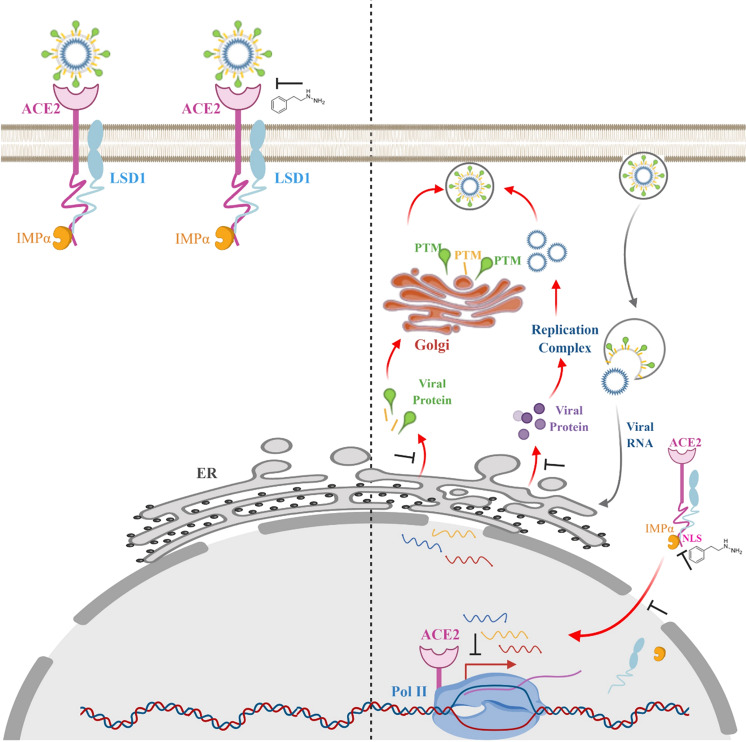
Fig. 6. A summary model of SARS-CoV-2/ACE2/LSD1 interactions.
Our data support a model in which: (Left) at the cell surface, SARS-CoV-2 entry relies on LSD1-dependent demethylation of ACE2, which is required for ACE2/SARS-CoV-2 spike binding. LSD1 inhibition therefore reduces viral entry via ACE2, especially LSD1 catalytic inhibitors such as GSK. In addition, our novel ACE2 peptide inhibitors, which compete with binding to SARS-CoV-2 spike protein, also block viral replication. (Right) Intracellularly, NACE2i blocks LSD1/ACE2 binding at cytoplasmic tail, and this change in protein–protein interactions might consequently inhibit LSD1’s demethylation activity. IMPα binds to the cytoplasmic tail of non-methylated ACE2 to mediate nuclear translocation and Pol II association to regulate gene expression. Therefore, NACE2i blocking IMPα/ACE2 binding impact ACE2-Pol II nuclear co-localization, which is required for SARS-CoV-2-related transcription, to reduce viral replication.