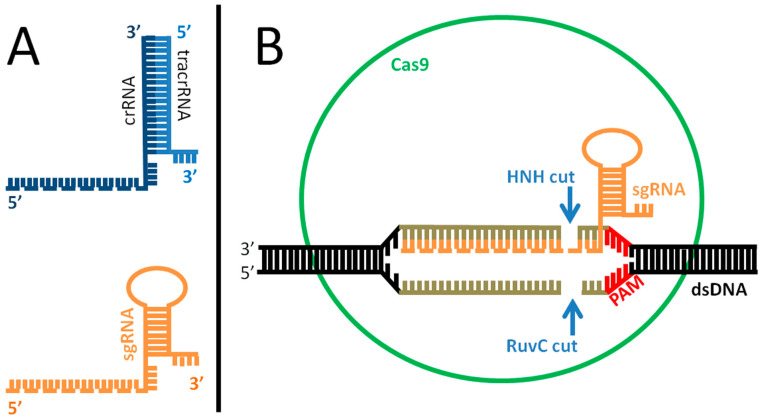
Figure 2.
Mechanism of action of the Cas complex. (A): difference between natural (top) and engineered (bottom) guide RNA (gRNA). The natural system is composed of two parts, crRNA (dark blue) and tracrRNA (light blue), which are paired and drive Cas9 to target the invading viral DNA. The portion of the crRNA recognizing the target is indicated as a dotted line. In the engineered form, the synthetic, single guide RNA (sgRNA) is one molecule that mimics the shape of its natural counterpart, including the target recognition site (dotted line). (B): for genome editing, the sgRNA (orange) is incorporated inside the Cas9 protein (green) and recognizes the double-stranded target DNA (black-grey) promoting the pairing. In case of homology (pairing sequence length: 21–72 bp, indicated in gray) and in the presence of a PAM sequence (red) on the target DNA, the Cas protein cuts the DNA 3 bp upstream of PAM, causing a double strand break, thus inactivating target gene function. The repair of the target chromosome damage in eukaryotic cells usually employs the error prone NHEJ (non-homologous end joining) mechanism; however, in presence of an exogenous DNA template (injected with the Cas complex and sgRNA), the cell may fix the break through homologous-driven repair, thus introducing the sequence of interest inside the genome.