Figure 2.
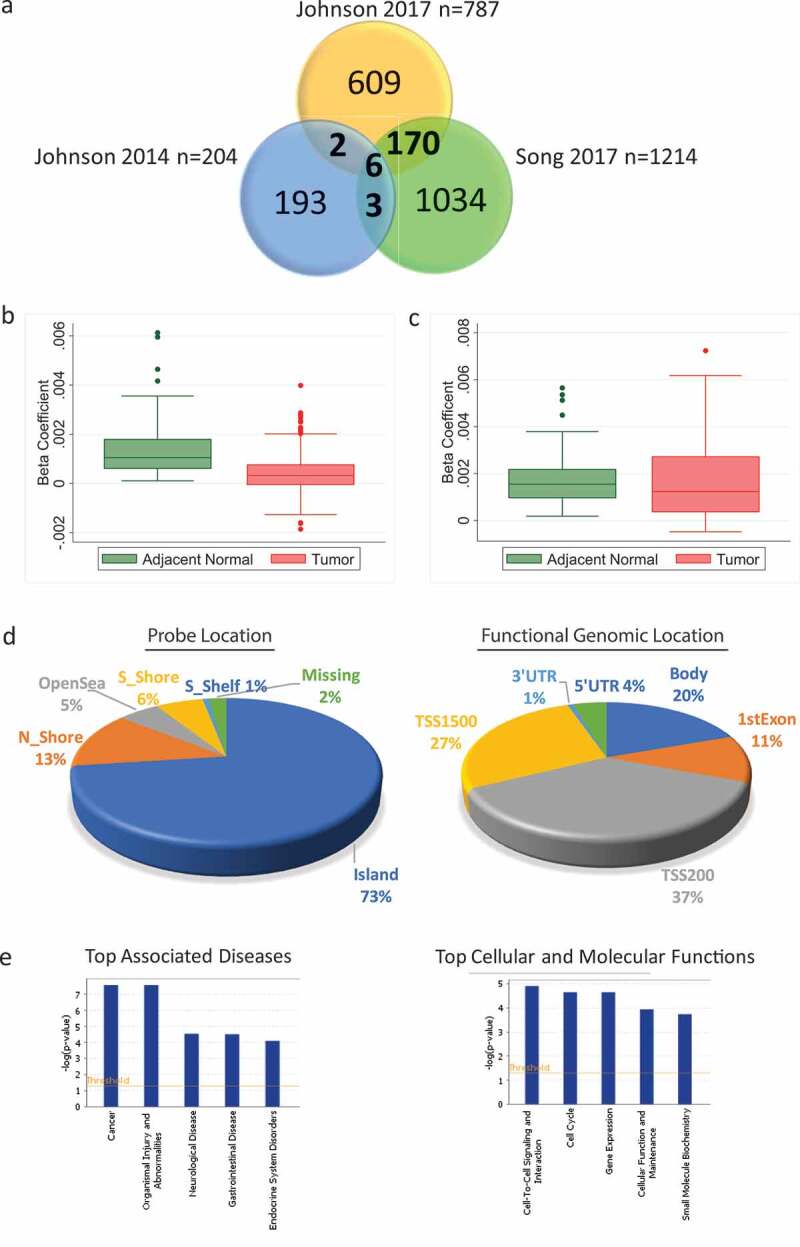
Consistent age-related methylation events across different studies in non-diseased breast tissue and in the adjacent normal tissue of breast cancer patients from Hong Kong (HK) and The Cancer Genome Atlas (TCGA). (a) Venn diagram comparing the number of age-related methylation events in three previously published genome-wide studies. (b) Box plot of beta coefficient values for both adjacent normal and tumour tissue for the 118 probes with increased age-related methylation identified in the HK study (n = 84). (c) Box plot of beta coefficient values for both adjacent normal and tumour tissue for the 105 probes with increased age-related methylation in TCGA (n = 82) (d) Pie chart with the location (left) and functional genomic location (right) of statistically significant age-related methylation probes from the HK study. (e) Ingenuity Pathway Analysis of 112 genes corresponding to probes with increased age-related methylation sites in the HK cohort
