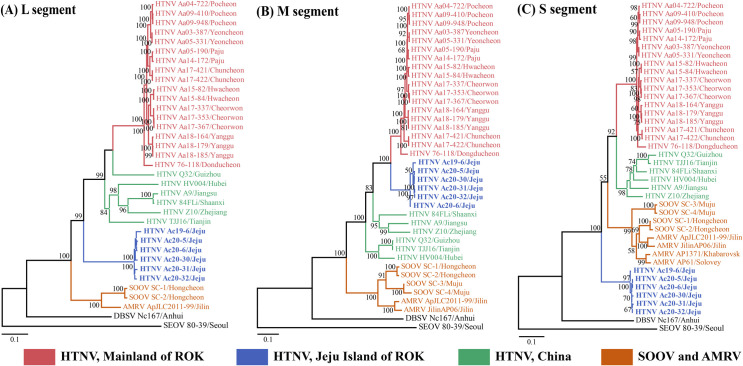
Fig 3. Phylogeographic analysis of the L, M, and S segments of Hantaan virus (HTNV) collected from rodents in Jeju Island, Republic of Korea (ROK).
Whole-genome sequences of HTNV from A. chejuensis of Jeju Island were obtained by multiplex polymerase chain reaction-based next-generation sequencing during 2018–2020. The phylogenies were generated by maximum likelihood methods in MEGA 7 with bootstrap 1,000 iterations based on the HTNV (A) L (1–6,533 nt), (B) M (1–3,618 nt), and (C) S (1–1,696 nt) segments, respectively. The branch lengths were proportional to the number of nucleotide substitutions, while vertical distances were used for clarity. The numbers at each node are bootstrap probabilities determined for 1,000 iterations. HTNV groups are color-coded according to geographic sites (red, mainland ROK; blue, Jeju Island of ROK; green, China). The brown color indicates SOOV and AMRV strains from East Asia (China, Russia, and ROK). The following hantaviral sequences were included in the analysis: HTNV Aa03-387 (L segment, KT934958; M segment, KT934992; S segment, KT935026), Aa04-722 (L segment, KU2071740; M segment, KU207182; S segment, KU207190), Aa05-190 (L segment, KT934959; M segment, KT934993; S segment, KT935027), Aa05-331 (L segment, KT934962; M segment, KT934996; S segment, KT935030), Aa09-410 (L segment, KU207177; M segment, KU207185; S segment, KU207193), Aa09-948 (L segment, KT934966; M segment, KT935000; S segment, KT935034), Aa14-172 (L segment, KT934974; M segment, KT935008; S segment, KT935042), Aa15-82 (L segment, MT012572; M segment, MT012560; S segment, MT012548), Aa15-84 (L segment, MT012573; M segment, MT012561; S segment, MT012549), Aa17-337 (L segment, MT012574; M segment, MT012562; S segment, MT012550), Aa17-353 (L segment, MT012575; M segment, MT012563; S segment, MT012551), Aa17-367 (L segment, MT012576; M segment, MT012564; S segment, MT012552), Aa17-421 (L segment, MT012577; M segment, MT012565; S segment, MT012553), Aa17-422 (L segment, MT012578; M segment, MT012566; S segment, MT012554), Aa18-164 (L segment, MT012579; M segment, MT012567; S segment, MT012555), Aa18-179 (L segment, MT012580; M segment, MT012568; S segment, MT012556), Aa18-185 (L segment, MT012581; M segment, MT012569; S segment, MT012557), Ac19-6 (L segment, MW219756; M segment, MW219762; S segment, MW219768), Ac20-5 (L segment, MW219757; M segment, MW219763; S segment, MW219769), Ac20-6 (L segment, MW219758; M segment, MW219764; S segment, MW219770), Ac20-30 (L segment, MW219759; M segment, MW219765; S segment, MW219771), Ac20-31 (L segment, MW219760; M segment, MW219766; S segment, MW219772), Ac20-32 (L segment, MW219761; M segment, MW219767; S segment, MW219773), 76–118 (L segment, NC_005222; M segment, M14627; S segment, M14626), HV004 (L segment, JQ083393; M segment, JQ083394; S segment, JQ093395), A9 (L segment, AF293665; M segment, AF035831; S segment, AF329390), Q32 (L segment, DQ371906; M segment, DQ371905; S segment, AB027097), 84FLi (L segment, AF336826; M segment, AF345636; S segment, AF366568), Z10 (L segment,NC_006435; M segment, NC_006437; S segment, AF366568), TJJ16 (L segment, KU215675; M segment, EU074672; S segment, AY839871), SOOV SC-1 (L segment, DQ056292; M segment, AY675353; S segment, AY675349), SC-2 (L segment, AY675354; M segment, DQ056293; S segment, AY675350), SC-3 (M segment, DQ056294; S segment, AY675351), SC-4 (M segment, DQ056295; S segment, AY675352), AMRV strains ApJLC2011-99 (L segment, JX473002; M segment, JX473003; S segment, JX473004), JilinAP06 (M segment, EF371454; S segment, EF121324), AP1371 (S segment, AF427324), AP61 (S segment, AB071183), DBSV Nc167 (L segment, DQ989237; M segment, AB027115; S segment, AB027523), and SEOV 80–39 (L segment, NC_005238; M segment, NC_005237; S segment, NC_005236). Abbreviations: SOOV, Soochong virus; AMRV, Amur virus; DBSV, Dàbiéshān virus; SEOV, Seoul virus.