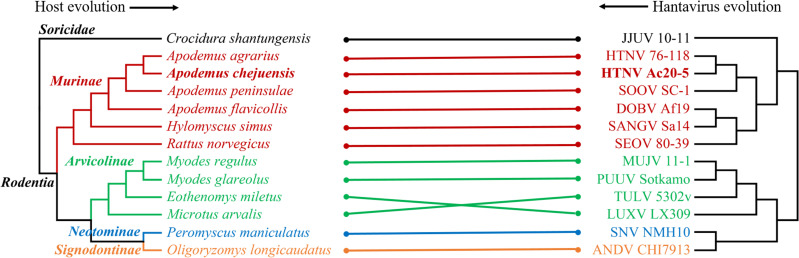
Fig 5. Tanglegram comparing the phylogenies between hantaviruses and their reservoir hosts in Rodentia.
Tanglegram was generated using the R package, using consensus maximum likelihood topologies based on the nucleotide sequences of the hantaviral M segment (right panel) with mitochondrial DNA cytochrome b gene sequences of the reservoir host species (left panel). Letters for taxa are shown in red for Murinae, green for Arvicolinae, blue for Signodontinae, orange for Neotominae, black for Soricidae in left panel. Apodemus chejuensis-borne HTNV (Ac20-5) is shown in bold lettering in each panel. The host species and viruses accession number followed up; reservoir hosts Crocidura shantungensis (HQ993053), A. agrarius (MN205319), A. chejuensis (MW219775), A. peninsulae (AB073811), A. flavicollis (JF819967), Hylomyscus simus (DQ212188), Rattus norvegicus (AB355903), Myodes regulus (NC_016427), Myodes glareolus (AF119272), Eothenomys miletus (AY426686), Microtus arvalis (EU439459), Peromyscus maniculatus (AF119261), Oligoryzomys longicaudatus (AF346566), hantavirus strains JJUV 10–11 (NC_034404), HTNV 76–118 (M14627), HTNV Ac20-5 (MW219763), SOOV SC-1 (AY675353), DOBV Af19 (NC_005234), SANGV Sa14 (NC_034516), SEOV 80–39 (NC_005237), MUJV 11–1 (JX028272), PUUV Sotkamo (HE801634), TULV 5302v (NC_005228), LUXV LX3009 (NC_038528), SNV NMH10 (NC_005215), ANDV CHI7913 (AY228238). JJUV, Jeju virus; HTNV, Hantaan virus; SOOV, Soochong virus; DOBV, Dobrava virus; SANGV, Sangassou virus; SEOV, Seoul virus; MUJV, Muju virus; PUUV, Puumala virus; TULV, Tula virus; LUXV, Lúxī virus; SNV, Sin Nombre virus; ANDV, Andes virus.