Figure 3.
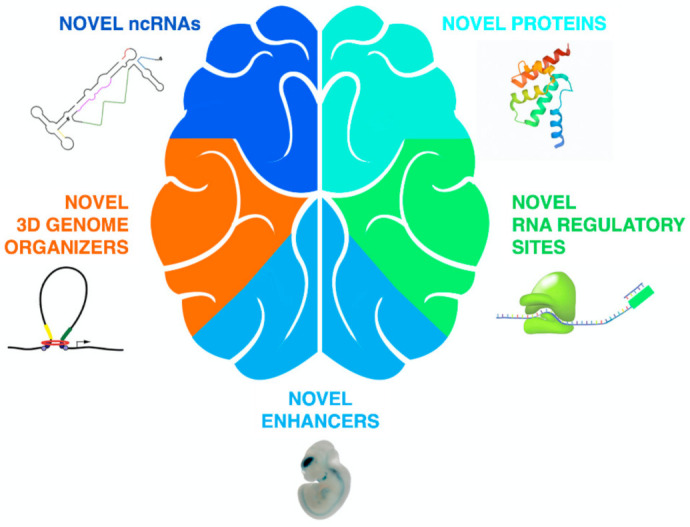
Summary of retrotransposon exaptations involved in brain evolution. Schematically recapitulated in the Figure are five major types of genomic novelty provided by RE exaptation that have been recognized to strongly affect brain evolution. Going clockwise from the left upper sector: REs can be a source of novel retrotransposon-derived ncRNAs (e.g., SINE-derived BC200 RNA); REs can be the source of novel proteins derived from retrotransposon (especially ERV) coding sequences; exonization of retrotransposon sequences (resulting in mRNAs or lncRNAs with embedded RE-derived sequences) can lead, for example, to the appearance of a new regulatory site (green box) within mRNA sequences, which can recruit miRNA, thereby affecting mRNA translation/stability; REs can provide new transcription TF binding sites, contributing to the birth of novel enhancers and altering the transcription of specific brain genes; REs can play a major role as 3D genome organizers, thereby strongly influencing the expression of brain-specific genes via changes in the 3D nuclear chromatin folding.
