Fig. 1.
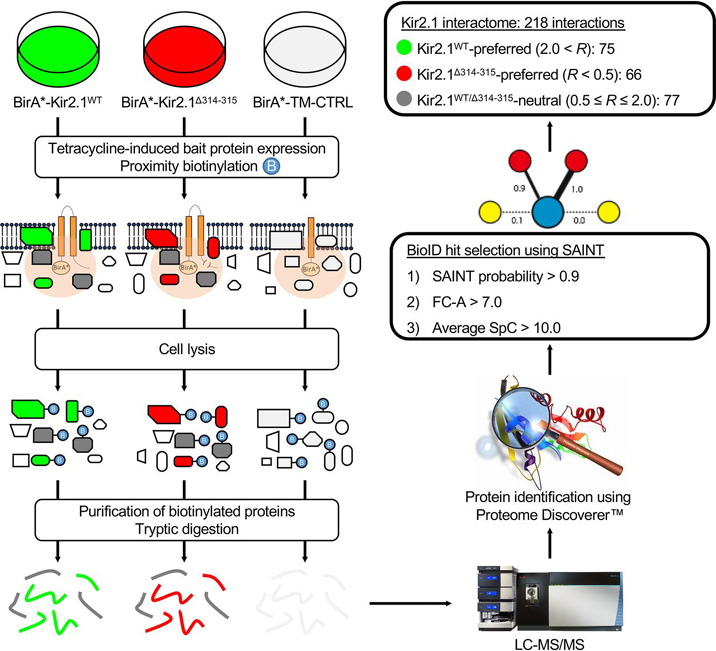
Overall procedure to generate the Kir2.1 BioID interactome map. Stable cells expressing BirA*-tagged Kir2.1WT, Kir2.1Δ314-315 or TM-CTRL bait proteins were generated using the Flp-In T-Rex 293 cell line. Expression of the bait proteins was induced by tetracycline and cells were treated with Supplemental biotin for 24 h. After cell lysis, biotinylated proteins were purified on streptavidin-agarose beads and digested with trypsin. Tryptic peptides were analyzed using LC–MS/MS and proteins were identified using Proteome Discoverer. After applying a stringent set of criteria, we identified 218 high-confidence Kir2.1 BioID hits. Using the normalized Kir2.1WT/Δ314-315 SpC ratio “R ”, we classified the interactors in three categories: 75 Kir2.1WT-preferred interactors, 66 Kir2.1Δ314-315-preferred interactors and 77 Kir2.1WT/Δ314-315-neutral interactors. CTRL: control; LC–MS/MS: liquid chromatography with tandem MS; SAINT: Significance Analysis of INTeractome; SpC: spectral counts; TM: transmembrane; WT: WT.
