Abstract
This updated systematic review and meta-analysis follows two aims: 1) to assess Mycobacterium tuberculosis (M. tuberculosis) antibiotic resistance in Iran from 2013 to 2020 and, 2) to assess the trend of resistance from 1999 to 2020. Several national and international databases were systematically searched through MeSH extracted keywords to identify 41 published studies addressing drug-resistant M. tuberculosis in Iran. Meta-analysis was done based on the PRISMA protocols using Comprehensive Meta-Analysis software. The average prevalence of resistance to first- and second-line anti-TB drugs, multidrug-resistant TB (MDR-TB) and extensively drug-resistant TB (XDR-TB) in new and previously treated tuberculosis (TB) cases in Iran during 2013–2020 were as follows: isoniazid 6.9%, rifampin 7.9%, ethambutol 5.7%, pyrazinamide 20.4%, para-aminosalicylic acid 4.6%, capreomycin 1.7%, cycloserine 1.8%, ethionamide 11.3%, ofloxacin 1.5%, kanamycin 3.8%, amikacin 2.2%, MDR-TB 6.3% and XDR-TB 0.9%. Based on the presented data, M. tuberculosis resistance to first- and second-line anti-TB drugs, as well as MDR-TB, was low during 2013–2020 in Iran. Furthermore, there was a declining trend in TB drug resistance from 1999 to 2020. Hence, to maintain the current decreasing trend and to control and eliminate TB infection in Iran, continuous monitoring of resistance patterns is recommended.
Key Words: Antibiotic, Iran, Meta-analysis, Mycobacterium tuberculosis, Resistance
Introduction
On 24 March 1882, Dr Robert Koch discovered a rod-shaped, aerobic bacterium called Mycobacterium tuberculosis (M. tuberculosis). It is the causative agent for tuberculosis (TB) which is one of the ten most common causes of death worldwide. The latent form of tubercle bacilli has been detected in 1.7 billion people around the world (1-3). Between 5 and 10% of these people will eventually develop the active form of the disease (pulmonary or extrapulmonary) in their lifetime (4, 5). TB is an air-borne and communicable disease and humans are the only natural reservoir of the infection (6). According to the latest report of the World Health Organization (WHO) in 2018, TB remained a global health problem with 10 million infected people (57% adult men, 32% adult women, and 11% children), 1.2 million deaths in HIV-negative and 251,000 deaths in HIV-positive patients (1). In 2014, WHO announced “the End TB Strategy” which aimed to reduce TB-associated deaths (by 90%) and incidence rate (by 80%) and to end the global TB epidemic between 2016 and 2035 (1). To achieve these purposes, developing novel diagnostic tests for rapid detection, use of effective drugs or new treatments, and appropriate vaccination are needed (1). Since 1921, the bacilli Calmette-Guérin (BCG) live attenuated vaccine has been the only approved vaccine for TB prevention in newborns and children. It has variable protection against adult pulmonary TB (0–80%) while it is unable to prevent the reactivation of latent TB (7, 8). Treatment with first-line drugs i. e., isoniazid, rifampin, ethambutol, and pyrazinamide is recommended for a six-month period in drug-susceptible TB as the mortality rate is high without treatment (1). The success rate for first-line drugs is >85%, however, the emergence of drug-resistant M. tuberculosis, particularly multidrug-resistant TB (MDR-TB defined as resistance to isoniazid and rifampin) has led to a decrease in treatment success to 56%, an increase in treatment duration and costs, and administration of more toxic second-line drugs (1). TB incidence in 2018 in Iran, a country with a population of 82 million located in the Middle East, showed a decreasing trend (11, 000; 14 cases per 100,000 population), however, it has shared geographical borders with high incidence countries such as Pakistan (6% in 2018) (1). Hence, continuous surveillance on TB epidemiology especially drug resistance status is of high necessity for disease control. M. tuberculosis antibiotic resistance pattern has previously been studied between March 1999 and May 2013 in Iran (9). Nevertheless, as the prevalence of TB antibiotic resistance is changing over time, we have performed the current systematic review and meta-analysis from 2013 to 2020 to update the evidence in Iran.
Methods
Search strategies and data sources
The current systematic review and meta-analysis was conducted based on the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analysis) guidelines (10). The computer-assisted systematic search was done in national and international databases including Scientific Information Database (SID) (www.sid.ir), Magiran (www.Magiran.com), ISI Web of Knowledge, PubMed, and Scopus. Each cross-sectional study published between May 2013 and March 30, 2020, in English or Persian languages on M. tuberculosis antibiotic resistance in Iran was investigated. M. tuberculosis, antibiotic resistance, and Iran were the main medical subject heading (MeSH) extracted keywords used for searching. The reference lists of all included articles were further assessed to find any missed relevant studies.
Data selection criteria and quality assessment
The titles, abstracts, and full texts of collected articles were further evaluated in detail to select eligible studies based on our inclusion and exclusion criteria. In addition, the Joanna Briggs Institute (JBI) critical appraisal checklist was selected to assess the quality of each included article. Eligible studies showed high quality (>5 scores), medium quality (4-5 scores), or low quality (<4 scores) (11). Original articles assessing the prevalence of M. tuberculosis antibiotic resistance with the following features were included in the study: (1) published in English or Persian, 2) limited to Iran, 3) clinical strains of M. tuberculosis isolated from new or previously treated cases, 4) studies evaluating monoresistance (which is defined as resistance to only 1 first- or second-line anti-TB drugs), MDR and XDR (extensively drug-resistant) status and 5) full-text availability. The following studies were excluded from analysis: 1) studies reporting drug resistance patterns in nontuberculous mycobacteria (NTM), 2) studies that only focused on drug resistance mechanisms, 3) studies that solely evaluated any drug resistance status (which is defined as resistance to any drug regardless of monoresistance), 4) non-original studies, 5) duplicate publications including same studies published in both English and Persian languages and two or more relevant studies conducted by the first or corresponding author in the same year and also several multicenter studies with identical information such as same authors’ name, same enrollment time and previously determined antibiotic resistance profiles, 6) studies available only in abstract form and dissertations, 7) studies without sufficient data such as study date or ambiguous information, 8) studies performed merely to evaluate the sensitivity and specificity of different antimicrobial susceptibility testing methods using previous banks of bacterial isolates or clinical isolates of M. tuberculosis with known drug resistance profiles, 9) studies on M. tuberculosis other than antibiotic resistance such as epidemiological patterns or treatment of the disease, and 10) multicenter studies without sufficient and separate data for each province.
Data extraction
Extracted information from eligible studies included in the meta-analysis is shown in Table 1. Important data were as follows: first author’s surname, publication date, region of study, study enrollment date, number of tested isolates, methods used for assessing bacterial antibiotic susceptibility, and prevalence of M. tuberculosis resistance to first- and second-line drugs.
Table 1.
Extracted information from eligible studies included in the meta-analysis during 2013-2020
| Author (Ref) |
Published date |
Province | Enrollment date | Strain (n) |
DST | Antibiotic resistance (n) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First-line drugs | Second-line drugs | |||||||||||||||||||||
| INH | RIF | EMB | PZA | STR | PAS | CAP | CYC | ETO | OFX | KAN | AMK | MDR | XDR | |||||||||
| Moradi et al (11) | 2017 | Ardabil | 2014-2015 | 9 | Agar proportional | 1 | 0 | ND | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Atashi et al (12) | 2017 | Ardabil | 2014 | 14 | Agar proportional | 2 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Sahebi et al (13) | 2016 | Ardabil | 2014 | 9 | Molecular | 3 | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Sahebi et al (14) | 2015 | Ardabil | 2011-2013 | 26 | Molecular | 3 | 4 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Velayati et al (15) | 2014 | Ardabil | 2010-2011 | 65 | Molecular | 2 | 4 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 4 | ND | |||
| Moradi et al (11) | 2017 | East Azerbaijan | 2014-2015 | 21 | Agar proportional | 2 | 2 | ND | 6 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Atashi et al (12) | 2017 | East Azerbaijan | 2014 | 28 | Agar proportional | 0 | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Sahebi et al (13) | 2016 | East Azerbaijan | 2014 | 28 | Molecular | 1 | 4 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Rashedi et al (16) | 2015 | East Azerbaijan | 2012-2014 | 48 | Agar proportional | 2 | 4 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 1 | ND | |||
| Sahebi et al (14) | 2015 | East Azerbaijan | 2011-2013 | 87 | Molecular | 3 | 15 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Moaddab et al (17) | 2015 | East Azerbaijan | 2009-2012 | 100 | Agar proportional | 4 | 5 | 1 | 18 | 21 | ND | ND | ND | ND | ND | ND | ND | 21 | ND | |||
| Amini et al (18) | 2019 | Fars | 2015-2017 | 19 | Agar proportional | 3 | 1 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 1 | ND | |||
| Zarei et al (19) | 2017 | Fars | 2012-2014 | 199 | Agar proportional | 38 | 30 | 16 | ND | 21 | ND | ND | ND | ND | ND | ND | ND | 22 | ND | |||
| Honarvar et al (20) | 2015 | Fars | 2012-2013 | 92 | Agar proportional | 16 | 19 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 24 | ND | |||
| Motamedifar et al (21) | 2015 | Fars | 2004-2013 | 59 | Agar proportional | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 6 | ND | |||
| Velayati et al (15) | 2014 | Fars | 2010-2011 | 40 | Molecular | 2 | 5 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 5 | ND | |||
| Mansoori et al (22) | 2018 | Golestan | 2014-2015 | 164 | Agar proportional | 3 | 0 | 0 | ND | 12 | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Velayati et al (15) | 2014 | Golestan | 2010-2011 | 47 | Molecular | 3 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 2 | ND | |||
| Velayati et al (15) | 2014 | Guilan | 2010-2011 | 39 | Molecular | 1 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 3 | ND | |||
| Moradi et al (11) | 2017 | Hamadan | 2014-2015 | 5 | Agar proportional | 0 | 0 | ND | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Atashi et al (12) | 2017 | Hamadan | 2014 | 11 | Agar proportional | 1 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Velayati et al (15) | 2014 | Hamadan | 2010-2011 | 21 | Molecular | 1 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0 | ND | |||
| Zamani et al (23) | 2016 | Hormozgan | 2012-2013 | 38 | Agar proportional | 4 | ND | ND | ND | 2 | ND | ND | ND | ND | ND | ND | ND | 3 | ND | |||
| Nasiri et al (24) | 2014 | Hormozgan | 2010-2012 | 48 | Agar proportional | 3 | 2 | 2 | ND | 4 | ND | ND | ND | ND | ND | ND | ND | 2 | ND | |||
| Velayati et al (15) | 2014 | Hormozgan | 2010-2011 | 38 | Molecular | 3 | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 3 | ND | |||
| Moradi et al (11) | 2017 | Ilam | 2014-2015 | 4 | Agar proportional | 0 | 0 | ND | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Atashi et al (12) | 2017 | Ilam | 2014 | 6 | Agar proportional | 1 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Amini et al (18) | 2019 | Isfahan | 2015-2017 | 19 | Agar proportional | 2 | 1 | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 1 | ND | |||
| Karimi et al (25) | 2017 | Isfahan | 2014-2015 | 205 | Agar proportional | 6 | 4 | 3 | ND | 10 | ND | ND | ND | ND | ND | ND | ND | 4 | ND | |||
| Nasr Esfahani et al (26) | 2016 | Isfahan | 2013 | 32 | Agar proportional | ND | ND | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Nasiri et al (24) | 2014 | Isfahan | 2010-2012 | 45 | Agar proportional | 2 | 2 | 0 | ND | 1 | ND | ND | ND | ND | ND | ND | ND | 2 | ND | |||
| Velayati et al (15) | 2014 | Isfahan | 2010-2011 | 42 | Molecular | 5 | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 2 | ND | |||
| Velayati et al (15) | 2014 | Kerman | 2010-2011 | 24 | Molecular | 1 | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 3 | ND | |||
| Mohammadi et al (27) | 2018 | Kermanshah | 2014-2015 | 50 | Agar proportional | ND | ND | 7 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 8 | ND | |||
| Moradi et al (11) | 2017 | Kermanshah | 2014-2015 | 17 | Agar proportional | 1 | 2 | ND | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Atashi et al (12) | 2017 | Kermanshah | 2014 | 31 | Agar proportional | 1 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Sahebi et al (13) | 2016 | Kermanshah | 2014 | 16 | Molecular | 1 | 4 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Sahebi et al (14) | 2015 | Kermanshah | 2011-2013 | 51 | Molecular | 1 | 5 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Mohajeri et al (28) | 2014 | Kermanshah | 2011-2012 | 112 | Agar proportional | 18 | 16 | 15 | 27 | 25 | 19 | ND | 4 | 14 | ND | ND | ND | 16 | ND | |||
| Nasiri et al (24) | 2014 | Kermanshah | 2010-2012 | 15 | Agar proportional | 4 | 3 | 3 | ND | 3 | ND | ND | ND | ND | ND | ND | ND | 3 | ND | |||
| Velayati et al (15) | 2014 | Kermanshah | 2010-2011 | 16 | Molecular | 1 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 1 | ND | |||
| Khosravi et al (29) | 2019 | Khuzestan | 2016-2017 | 307 | Agar proportional | 6 | 10 | 10 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 4 | ND | |||
| Khosravi et al (30) | 2019 | Khuzestan | 2015-2017 | 37 | Agar proportional | 3 | 16 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 5 | ND | |||
| Amini et al (18) | 2019 | Khuzestan | 2015-2017 | 20 | Agar proportional | 1 | 1 | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 1 | ND | |||
| Badie et al (31) | 2016 | Khuzestan | 2015 | 64 | Agar proportional | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 2 | ND | |||
| Khosravi et al (32) | 2017 | Khuzestan | 2013-2014 | 88 | Agar proportional | 41 | 35 | 5 | ND | 32 | ND | ND | ND | ND | ND | ND | ND | 22 | ND | |||
| Velayati et al (15) | 2014 | Khuzestan | 2010-2011 | 119 | Molecular | 7 | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 6 | ND | |||
| Khosravi et al (33) | 2014 | Khuzestan | 2010-2011 | 160 | Molecular | 18 | 20 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 8 | ND | |||
| Moradi et al (11) | 2017 | Kurdistan | 2014-2015 | 27 | Agar proportional | 0 | 0 | ND | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Atashi et al (12) | 2017 | Kurdistan | 2014 | 23 | Agar proportional | 0 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Sahebi et al (13) | 2016 | Kurdistan | 2014 | 12 | Molecular | 3 | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Sahebi et al (14) | 2015 | Kurdistan | 2011-2013 | 50 | Molecular | 3 | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Velayati et al (15) | 2014 | Kurdistan | 2010-2011 | 16 | Molecular | 2 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0 | ND | |||
| Heidary et al (34) | 2020 | Lorestan | 2014-2017 | 106 | Agar proportional | 1 | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 4 | ND | |||
| Moradi et al (11) | 2017 | Lorestan | 2014-2015 | 19 | Agar proportional | 0 | 0 | ND | 4 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Atashi et al (12) | 2017 | Lorestan | 2014 | 27 | Agar proportional | 0 | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Velayati et al (15) | 2014 | Lorestan | 2010-2011 | 24 | Molecular | 0 | 5 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0 | ND | |||
| Farazi et al (35) | 2013 | Markazi | 2011-2012 | 115 | Agar proportional | 3 | 2 | 8 | ND | 3 | ND | ND | ND | ND | ND | ND | ND | 9 | ND | |||
| Velayati et al (15) | 2014 | Markazi | 2010-2011 | 15 | Molecular | 3 | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 2 | ND | |||
| Babamahmoodi et al (36) | 2014 | Mazandaran | 2013 | 54 | Molecular | 2 | 3 | ND | ND | 4 | ND | ND | ND | ND | ND | 3 | 3 | ND | ND | |||
| Velayati et al (15) | 2014 | Mazandaran | 2010-2011 | 26 | Molecular | 1 | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 1 | ND | |||
| Atashi et al (12) | 2017 | Qazvin | 2014 | 5 | Agar proportional | 0 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Velayati et al (15) | 2014 | Qazvin | 2010-2011 | 10 | Molecular | 1 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 2 | ND | |||
| Velayati et al (15) | 2014 | Qom | 2010-2011 | 61 | Molecular | 3 | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 4 | ND | |||
| Amini et al (18) | 2019 | Razavi Khorasan | 2015-2017 | 56 | Agar proportional | 4 | 5 | 8 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 4 | ND | |||
| Sani et al (37) | 2015 | Razavi Khorasan | 2012-2013 | 100 | Agar proportional | 7 | 7 | 3 | ND | 9 | ND | ND | ND | ND | ND | ND | ND | 4 | ND | |||
| Danesh et al (38) | 2014 | Razavi Khorasan | 2011-2012 | 48 | Agar proportional | 0 | 0 | 0 | 0 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Velayati et al (15) | 2014 | Razavi Khorasan | 2010-2011 | 117 | Molecular | 10 | 9 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 2 | ND | |||
| Velayati et al (15) | 2014 | Semnan | 2010-2011 | 21 | Molecular | 0 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0 | ND | |||
| Hashemi Shahri et al (39) | 2019 | Sistan and Balouchastan | 2013-2016 | 100 | Agar proportional | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 2 | ND | |||
| Shirazinia et al (40) | 2017 | Sistan and Balouchastan | 2010-2013 | 525 | Agar proportional | 15 | 16 | 0 | ND | 0 | ND | ND | ND | ND | ND | ND | ND | 7 | ND | |||
| Nasiri et al (24) | 2014 | Sistan and Balouchastan | 2010-2012 | 59 | Agar proportional | 5 | 3 | 3 | ND | 8 | ND | ND | ND | ND | ND | ND | ND | 3 | ND | |||
| Velayati et al (15) | 2014 | Sistan and Balouchastan | 2010-2011 | 165 | Molecular | 8 | 10 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 1 | ND | |||
| Vaziri et al (41) | 2019 | Tehran | 2014-2018 | 606 | Agar proportional | 3 | 3 | 5 | ND | 2 | ND | ND | ND | ND | ND | ND | ND | 3 | 13 | |||
| Habibnia et al (42) | 2019 | Tehran | 2012-2018 | 100 | Agar proportional | 15 | 6 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 17 | ND | |||
| Aghajani et al (43) | 2019 | Tehran | 2011-2018 | 6937 | Molecular | 1617 | 1326 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 956 | ND | |||
| Amini et al (18) | 2019 | Tehran | 2015-2017 | 220 | Agar proportional | 11 | 4 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 11 | ND | |||
| Sakhaee et al (44) | 2017 | Tehran | 2013-2016 | 395 | Agar proportional | 24 | 24 | 40 | ND | 60 | ND | 12 | ND | ND | 12 | 7 | ND | 22 | 4 | |||
| Khanipour et al (45) | 2016 | Tehran | 2010-2015 | 723 | Agar proportional | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 15 | 8 | |||
| Imani Fooladi et al (46) | 2014 | Tehran | 2009-2011 | 103 | Agar proportional | 12 | 9 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 9 | ND | |||
| Tasbiti et al (47) | 2017 | Tehran | 2006-2014 | 1442 | Agar proportional | 168 | 176 | 169 | ND | 330 | 16 | 13 | 15 | 13 | 10 | 12 | 15 | 33 | 3 | |||
| Sharifipour (48) | 2014 | Tehran | 2011-2012 | 190 | Agar proportional | 12 | 5 | 8 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 30 | ND | |||
| Nasiri et al (24) | 2014 | Tehran | 2010-2012 | 85 | Agar proportional | 6 | 7 | 6 | ND | 14 | ND | ND | ND | ND | ND | ND | ND | 6 | ND | |||
| Bahrami et al (49) | 2013 | Tehran | 2010-2012 | 176 | Agar proportional | 12 | 19 | 48 | ND | ND | ND | ND | ND | ND | ND | ND | ND | 10 | ND | |||
| Pooideh et al (50) | 2015 | Tehran | 2010-2011 | 100 | Agar proportional | 13 | 23 | 26 | ND | 37 | ND | ND | ND | 61 | ND | 21 | ND | 4 | ND | |||
| Velayati et al (15) | 2014 | Tehran | 2010-2011 | 324 | Molecular | 20 | 26 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 32 | ND | |||
| Varahram et al (51) | 2014 | Tehran | 2003-2011 | 4825 | Agar proportional Molecular | 296 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Moradi et al (11) | 2017 | West Azerbaijan | 2014-2015 | 10 | Agar proportional | 0 | 0 | ND | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Atashi et al (12) | 2017 | West Azerbaijan | 2014 | 12 | Agar proportional | 0 | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Sahebi et al (13) | 2016 | West Azerbaijan | 2014 | 25 | Molecular | 1 | 4 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Sahebi et al (14) | 2015 | West Azerbaijan | 2011-2013 | 43 | Molecular | 1 | 3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| Velayati et al (15) | 2014 | Yazd | 2010-2011 | 12 | Molecular | 0 | 1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0 | ND | |||
| Atashi et al (12) | 2017 | Zanjan | 2014 | 5 | Agar proportional | 0 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
INH-isoniazid; RIF-rifampin; EMB-ethambutol; PZA-pyrazinamide; STR-streptomycin; PAS-para-aminosalicylic acid; CAP-capreomycin; CYC-cycloserine; ETO-ethionamide; OFX-ofloxacin; KAN-kanamycin; AMK-amikacin; MDR-multiple drug-resistant; XDR-extensively drug-resistant; DST-drug susceptibility testing; ND-not determined
Data analysis
Meta-analyses were conducted using the Comprehensive Meta-Analysis (CMA) (Biostat, Englewood, NJ) software package, and antimicrobial resistance trends were determined by the Graphpad Prism software package. Important analyzed parameters included the rates of M. tuberculosis antibiotic resistance and heterogeneity and publication bias among studies. The pooled estimates of the resistance prevalence for each drug were reported as a percentage and 95% confidence intervals (CIs) using a random-effects model (heterogeneity ≥25%;) or fixed-effects model (heterogeneity <25%;). The possibility of heterogeneity was evaluated by I² statistic and the Chi-square test with the Cochrane Q statistic. The existence of publication bias was assessed by Funnel plots.
Results
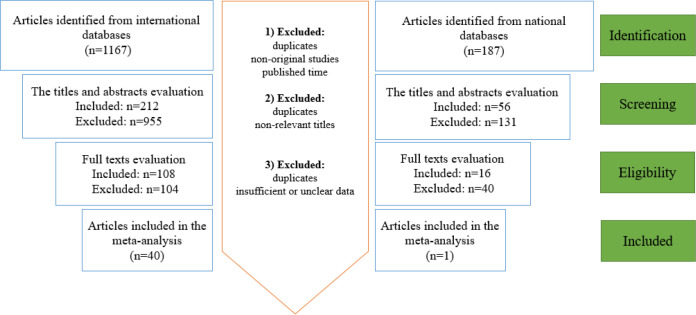
As shown in Figure 1, a total of 1,354 articles in Persian and English languages were collected during early literature search. At the end of the screening, 41 eligible articles were included in the meta-analysis based on the predefined inclusion and exclusion criteria. Forty-one included studies describing the prevalence of drug-resistant M. tuberculosis were selected from different provinces of Iran (Table 1). The most common laboratory methods that were used for assessing M. tuberculosis antibiotic susceptibility were agar proportion and molecular techniques such as line probe assay (LPA), real-time polymerase chain reaction (PCR), multiplex allele-specific polymerase chain reaction (MAS-PCR), and multiplex PCR.
Figure 1.
Summary of the literature search and study selection in the meta-analysis
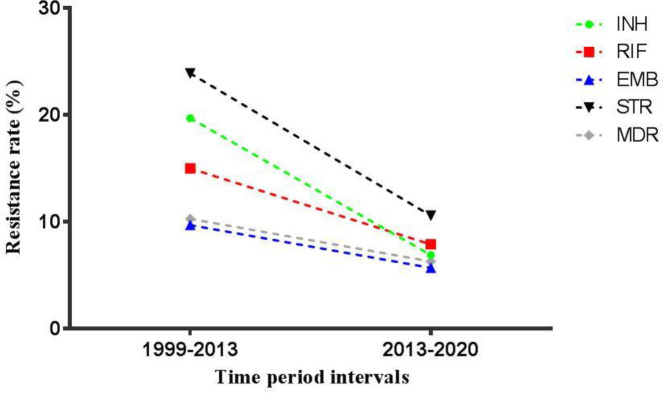
The total prevalence of isoniazid-resistant M. tuberculosis among new and previously treated cases in Iran was 6.9% (95% CI: 5.5-8.6) between 2013 and 2020, which was estimated using a random-effects model due to existence of significant heterogeneity (I2 = 92.1%; Q = 1101.5; P = 0.00). As shown in Table 2, the highest prevalence of isoniazid-resistant M. tuberculosis strains in different provinces of Iran was found in Fars (16.7%) and the lowest rates were seen in Semnan (0%), Yazd (0%), and Zanjan (0%). Additionally, Figure 2 demonstrates a decreasing trend in the incidence of isoniazid-resistant strains from 2013 to 2020 (6.9%) in comparison with 1999-2013 (19.7%) in Iran. According to the random-effects model (I2=83.7%; Q=516.4; P=0.00), the prevalence of rifampin-resistant M. tuberculosis strains among new and previously treated cases in Iran was 7.9% (95% CI: 6.6-9.5) between 2013 and 2020 which was lower than the previous report from 1999 to 2013 (15%). M. tuberculosis strains isolated from Fars (16.1%), Semnan (0%), and Zanjan (0%) showed the highest and lowest rates of resistance to rifampin, respectively. Among new and previously treated cases between 2013 and 2020, the mean resistance to ethambutol in Iran was 5.7% (95% CI: 4-8). The rate of resistance was highest in Kermanshah (14.2%) and lowest in Golestan (0%). The overall prevalence of ethambutol-resistant M. tuberculosis strains in Iran was calculated using the random-effects model (I2 = 86%; Q = 208.3; P = 0.00). As presented in Figure 2, ethambutol-resistant strains also showed a decreasing pattern in Iran from 1999 to 2020 (9.7% to 5.7%). Frequency of M. tuberculosis resistance to pyrazinamide calculated using the fixed-effects model was 20.4% (95% CI: 15.7-26.2; I2 = 13.2%; Q = 11.5; P = 0.31). The highest resistance rate was found in Hamadan (40%) and the lowest rate was seen in Razavi Khorasan (0%).
Table 2.
Mycobacterium tuberculosis antibiotic resistance profiles in different provinces of Iran during 2013-2020
| Province | Antibiotic resistance (%) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First-line drugs | Second-line drugs | |||||||||||||
| INH | RIF | EMB | PZA | STR | PAS | CAP | CYC | ETO | OFX | KAN | AMK | MDR | XDR | |
| Ardabil | 11.9 | 8.9 | ND | 11.1 | ND | ND | ND | ND | ND | ND | ND | ND | 6.1 | ND |
| East Azerbaijan | 4.3 | 9.9 | 2.4 | 20.1 | 21 | ND | ND | ND | ND | ND | ND | ND | 8.3 | ND |
| Fars | 16.7 | 16.1 | 7.8 | ND | 10.5 | ND | ND | ND | ND | ND | ND | ND | 13.6 | ND |
| Golestan | 3.4 | 1.5 | 0 | ND | 7.3 | ND | ND | ND | ND | ND | ND | ND | 4.2 | ND |
| Guilan | 2.5 | 5.1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 7.6 | ND |
| Hamadan | 6.9 | 8.1 | ND | 40 | ND | ND | ND | ND | ND | ND | ND | ND | 0 | ND |
| Hormozgan | 8.2 | 6.1 | 4.1 | ND | 7.1 | ND | ND | ND | ND | ND | ND | ND | 6.7 | ND |
| Ilam | 14 | 8.4 | ND | 25 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Isfahan | 6.2 | 3.7 | 2.7 | ND | 4.5 | ND | ND | ND | ND | ND | ND | ND | 3.2 | ND |
| Kerman | 4.1 | 12.5 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 12.5 | ND |
| Kermanshah | 9.4 | 13.8 | 14.2 | 22.9 | 22.1 | 16.9 | ND | 3.5 | 12.5 | ND | ND | ND | 14.2 | ND |
| Khuzestan | 8.9 | 11.7 | 4 | ND | 36.3 | ND | ND | ND | ND | ND | ND | ND | 6.1 | ND |
| Kurdistan | 8.7 | 6.1 | ND | 11.1 | ND | ND | ND | ND | ND | ND | ND | ND | 0 | ND |
| Lorestan | 1.5 | 4.5 | ND | 21 | ND | ND | ND | ND | ND | ND | ND | ND | 3.5 | ND |
| Markazi | 7.4 | 6.3 | 6.9 | ND | 2.6 | ND | ND | ND | ND | ND | ND | ND | 8.6 | ND |
| Mazandaran | 3.8 | 5.1 | ND | ND | 7.4 | ND | ND | ND | ND | ND | 5.5 | 5.5 | 3.8 | ND |
| Qazvin | 9.4 | 6.1 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 20 | ND |
| Qom | 4.9 | 4.9 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 6.5 | ND |
| Razavi Khorasan | 7.4 | 7.4 | 5 | 0 | 4.6 | ND | ND | ND | ND | ND | ND | ND | 4.1 | ND |
| Semnan | 0 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0 | ND |
| Sistan and Balouchastan | 4.1 | 4.3 | 0.9 | ND | 1.4 | ND | ND | ND | ND | ND | ND | ND | 1.9 | ND |
| Tehran | 7.6 | 7.4 | 7.6 | ND | 14.7 | 1.1 | 1.7 | 1 | 10.7 | 1.5 | 3.3 | 1 | 5.8 | 0.9 |
| West Azerbaijan | 3.4 | 10.2 | ND | 30 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| Yazd | 0 | 8.3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0 | ND |
| Zanjan | 0 | 0 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
INH-isoniazid; RIF-rifampin; EMB-ethambutol; PZA-pyrazinamide; STR-streptomycin; PAS-para-aminosalicylic acid; CAP-capreomycin; CYC-cycloserine; ETO-ethionamide; OFX-ofloxacin; KAN-kanamycin; AMK-amikacin; MDR-multiple drug-resistant; XDR-extensively drug-resistant; ND-not determined
Figure 2.
Antimicrobial resistance trends of Mycobacterium tuberculosis strains to isoniazid, rifampin, ethambutol, streptomycin, as well as MDR-TB, among new and previously treated cases in Iran from 1999 to 2020
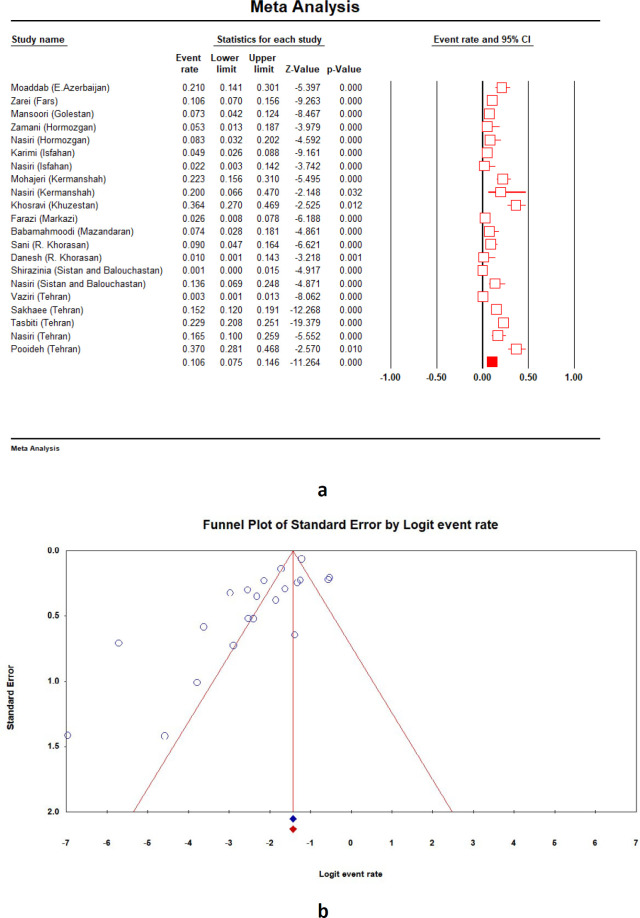
Second-line anti-TB drugs (groups A-D) used for MDR treatment include 1) group A: fluoroquinolones (ofloxacin, levofloxacin, moxifloxacin, and ciprofloxacin), 2) group B: injectable drugs (aminoglycosides (kanamycin, amikacin, and streptomycin) and cyclic peptides (capreomycin)), 3) group C: ethionamide/prothionamide, cycloserine/terizidone, linezolid and clofazimine, and 4) group D: D2 (bedaquiline and delamanid) and D3 ( para-aminosalicylic acid, imipenem-cilastatin, meropenem, amoxicillin/clavulanate and thioacetazone). The prevalence of M. tuberculosis resistance to these drugs which are mainly used in treatment of MDR-TB was not completely evaluated in the previous study conducted from 1999 to 2013 in Iran. Figure 3 shows the forest plot and funnel plot of streptomycin-resistant M. tuberculosis strains for new and previously treated cases between 2013 and 2020 in Iran. The prevalence of M. tuberculosis resistance to streptomycin was estimated at 10.6% (95% CI: 7.5-14.6; I2 = 89.6%; Q = 192.8; P = 0.00). The maximum and minimum resistance rates were observed in Khuzestan (36.3%) and Sistan and Balouchastan (1.4%), respectively. Similar to first-line anti-TB drugs, the trend of M. tuberculosis resistance to streptomycin decreased between 1999 and 2020 (23.9% to 10.6%) (Figure 2). Frequency of M. tuberculosis resistance to other drugs during 2013 to 2020 were as follows: para-aminosalicylic acid 4.6% (95% CI: 0.3-45.1; I2 = 98.4%; Q = 66.5; P = 0.00), capreomycin 1.7% (95% CI: 0.5-5.4; I2 = 89.3%; Q = 9.3; P = 0.00), cycloserine 1.8% (95% CI: 0.5-5.9; I2 = 79.4%; Q = 4.8; P = 0.02), ethionamide 11.3% (95% CI: 0.6-72.9; I2 = 99.1%; Q = 224.5; P = 0.00), ofloxacin 1.5% (95% CI: 0.3-6.1; I2 = 91.7%; Q = 12.0; P = 0.00), kanamycin 3.8% (95% CI: 0.6-20; I2 = 96.7%; Q = 91.4; P = 0.00), and amikacin 2.2% (95% CI: 0.4-10.9; I2 = 85.8%; Q = 7.0; P = 0.00).
Figure 3.
Forest plot (a) and funnel plot (b) showing streptomycin-resistant Mycobacterium tuberculosis prevalence among new and previously treated cases between 2013 and 2020 in Iran
Additionally, the pooled estimate of MDR- and XDR-TB in Iran were 6.3% (95% CI: 5-8; I2 = 86.7%; Q = 446.3; P = 0.00) and 0.9% (95% CI: 0.4-2.2; I2 = 78.3%; Q = 13.8; P = 0.00), respectively. Similar to first-line anti-TB drugs, the rate of MDR M. tuberculosis in Iran between 1999 and 2020 showed a downward trend (10.3% to 6.3%). The prevalence of MDR strains was highest in Qazvin (20%) and lowest in Hamadan, Kurdistan, Semnan, and Yazd (0%).
Discussion
Despite the strategies initiated by the National Tuberculosis Control Program (NTP) in 1996, TB continues to be a major health concern in Iran. One of the main reasons is the emergence of drug-resistant M. tuberculosis strains (53). The WHO implemented a general project back in 1994 in which data on anti-TB drug resistance are collected from numerous countries; the results indicate that the prevalence of drug-resistant M. tuberculosis strains is still a major public health concern around the world (1). The current meta-analysis is the newest report on the M. tuberculosis antibiotic resistance in Iran. We believe it provides such overviews of the available data to estimate the total antibiotic resistance. Also, it helps to increase our knowledge on drug-resistant TB trends during the years to prevent further rise in drug resistance and treatment failures. Among the first-line anti-TB drugs recommended by NTP to treat new sputum-positive TB cases in Iran, isoniazid and rifampin are the most effective agents (16). Isoniazid has also been recommended as a preventive therapy in latent TB infections (54). Therefore, isoniazid- and rifampin-resistant cases can increase the risk of treatment failure due to development of MDR-TB. Our analysis showed that 6.9% and 7.9% of both new and previously treated cases between 2013 and 2020 in Iran were resistant to isoniazid and rifampin, respectively. Based on the WHO report in 2018, the global average of isoniazid resistance varies between 7.2% among new TB cases and 11.6% in previously treated TB cases (1). Additionally, findings of the present study on the prevalence of rifampin-resistant strains of M. tuberculosis (7.9%) is comparable with international data from the Western Pacific Region (24%), European Region (10%), South-East Asian Region (6%), African Region (3%) and Region of America (1%) (55). In 2018, WHO announced that the incidence of MDR-TB and rifampicin-resistant TB in new and previously treated TB cases were 3.4% and 18% in the world, respectively (1). The rate of MDR-TB in Iran was low (6.3%). Patients with rifampicin-resistant TB and MDR-TB must be treated with second-line drugs (1). Only a few studies have investigated M. tuberculosis resistance to the second-line anti-TB drugs. This could be due to a low resistance rate to first-line anti-TB drugs in Iran. In addition to MDR-TB and rifampicin-resistant TB, global surveillance and treatment of XDR-TB (defined as MDR-TB plus resistance to at least one of the fluoroquinolones and one of the injectable agents) is completely urgent (1). A total of 28 XDR-TB cases were identified in Iran and 13,068 XDR-TB cases were reported in 2018 by WHO globally (1).
Among first-line anti-TB agents, the highest and lowest resistance rates between 2013 and 2020 in Iran were seen for pyrazinamide (20.4%) and ethambutol (5.7%). It has been suggested that chromosomal mutations are associated with development of drug resistance in clinical M. tuberculosis strains including rifampicin resistance-associated mutations in rpoB gene, mutations in katG or inhA genes in isoniazid-resistant strains, embB or ubiA genes mutations in ethambutol-resistant isolates, and mutations in pncA, rpsA, panD and clpC1 genes among pyrazinamide-resistant isolates (56). Similar resistance mechanisms were seen in M. tuberculosis isolated in Iran (data not shown).
Furthermore, the prevalence of resistant strains to isoniazid, rifampin, ethambutol, and streptomycin as well as MDR-TB in Iran showed a downward trend from 1999 to 2020 (Figure 2). This could be attributed to differences in the methods used for assessing drug susceptibility/antibiotic resistance due to possible heterogeneity among studies (fixed- or random-effects models) or the number of included studies among two reviews. Also, decreased incidence of TB cases in Iran (21 per 100,000 population in 2011 compared with 14 cases in 2018) and fewer Afghan refugees due to sanctions, are other contributory factors. The limitations that need to be acknowledged in this study include: 1) small number of studies reporting resistance to second-line drugs and XDR-TB, 2) lack of studies reporting M. tuberculosis drug resistance in some provinces, 3) lack of individual separation of drug-resistant TB in many studies according to new or previously treated patients, age, ethnicity, nationality (Iranian versus Afghans), and 4) existence of heterogeneity and publication bias among included studies.
Conclusion
The current updated systematic review and meta-analysis summarized the total prevalence of drug-resistant M. tuberculosis strains in new and previously treated TB cases (2013–2020) and drug-resistant TB trend (1999–2020) in Iran. Based on our results, the mean resistance to first- and second-line anti-TB drugs was low in Iran during 2013–2020 as were the cases of MDR-TB and XDR-T. There was also a decreasing trend in resistance of M. tuberculosis from 1999 to 2020. Hence, to continue the current downward trend and control and eliminate TB infections in Iran, continuous monitoring of resistance patterns through optimized and rapid diagnostic tests such as molecular techniques in all provinces of Iran is recommended.
Conflicts of Interest
The author declares that there are no conflicts of interest.
References
- 1.World Health Organization. Global tuberculosis report 2019. Gevena [Switzerland]: World Health Organization; 2019. [Google Scholar]
- 2.Murray PR, Rosenthal KS, Pfaller MA. Medical microbiology. 8th ed. UK: Elsevier Health Sciences: 2015. pp. 221–225. [Google Scholar]
- 3.Carroll KC, Butel JS, Morse SA. Jawetz Melnick & Adelbergs medical microbiology. 27th ed. Pennsylvania: McGraw Hill Professional; 2016. pp. 309–317. [Google Scholar]
- 4.Khademi F, Taheri RA, Avarvand AY, Vaez H, Momtazi-Borojeni AA, Soleimanpour S. Are chitosan natural polymers suitable as adjuvant/delivery system for anti-tuberculosis vaccines? Microb Pathog. 2018;121:218–223. doi: 10.1016/j.micpath.2018.05.035. [DOI] [PubMed] [Google Scholar]
- 5.Khademi F, Yousefi-Avarvand A, Derakhshan M, Najafi A, Tafaghodi M. Enhancing immunogenicity of novel multistage subunit vaccine of Mycobacterium tuberculosis using PLGA:DDA hybrid nanoparticles and MPLA: Subcutaneous administration. Iran J Basic Med Sci. 2019;22:893–900. doi: 10.22038/ijbms.2019.33962.8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khademi F, Taheri RA, Momtazi-Borojeni AA, Farnoosh G, Johnston TP, Sahebkar A. Potential of cationic liposomes as adjuvants/delivery systems for tuberculosis subunit vaccines. Rev Physiol Biochem Pharmacol. 2018:47–69. doi: 10.1007/112_2018_9. [DOI] [PubMed] [Google Scholar]
- 7.Yousefi-Avarvand A, Tafaghodi M, Soleimanpour S, Khademi F. HspX protein as a candidate vaccine against Mycobacterium tuberculosis: an overview. Front Biol. 2018;13:293–296. [Google Scholar]
- 8.Khademi F, Derakhshan M, Yousefi-Avarvand A, Tafaghodi M, Soleimanpour S. Multi-stage subunit vaccines against Mycobacterium tuberculosis: an alternative to the BCG vaccine or a BCG-prime boost? Expert Rev Vaccines. 2018;17:31–44. doi: 10.1080/14760584.2018.1406309. [DOI] [PubMed] [Google Scholar]
- 9.Nasiri MJ, Dabiri H, Darban-Sarokhalil D, Rezadehbashi M, Zamani S. Prevalence of drug-resistant tuberculosis in Iran: systematic review and meta-analysis. Am J Infect Control. 2014;42:1212–1218. doi: 10.1016/j.ajic.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 10.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6:e1000100. doi: 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munn Z, Moola S, Lisy K, Riitano D, Tufanaru C. Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and incidence data. Int J Evid Based Healthc. 2015;13:147–153. doi: 10.1097/XEB.0000000000000054. [DOI] [PubMed] [Google Scholar]
- 12.Moradi J, Mohajeri P, Alvandi A, Farahani A, Atashi S, Nasseri K. Molecular identification of mutations associated with pyrazinamide-resistance in multidrug-resistant tuberculosis in eight provinces of iran. J Clin Diagn Res. 2017;11:DC9–12. [Google Scholar]
- 13.Atashi S, Izadi B, Jalilian S, Madani SH, Farahani A, Mohajeri P. Evaluation of GeneXpert MTB/RIF for determination of rifampicin resistance among new tuberculosis cases in west and northwest Iran. New Microbes New Infect. 2017;19:117–20. doi: 10.1016/j.nmni.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahebi L, Ansarin K, Monfaredan A, Farajnia S, Nili S, Khalili M. Rapid detection of rifampicin-and isoniazid-resistant Mycobacterium tuberculosis using real-time PCR. Jundishapur J Microbiol. 2016;9:e29147. doi: 10.5812/jjm.29147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahebi L, Ansarin K, Farajnia S, Monfaredan A, Sabour S. Prevalence and risk factors of drug-resistant tuberculosis in border provinces of Iran. Postgrad Med. 2015;127:600–606. doi: 10.1080/00325481.2015.1069168. [DOI] [PubMed] [Google Scholar]
- 16.Velayati AA, Farnia P, Mozafari M, Sheikholeslami MF, Karahrudi MA, Tabarsi P, Hoffner S. High prevelance of rifampin-monoresistant tuberculosis: a retrospective analysis among Iranian pulmonary tuberculosis patients. Am J Trop Med Hyg. 2014;90:99–105. doi: 10.4269/ajtmh.13-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rashedi J, Mahdavi Poor B, Rafi A, Asgharzadeh M, Abdolalizadeh J, Moaddab SR. Multidrug-resistant tuberculosis in north-west of Iran and Republic of Azerbaijan: a major public health concern for Iranian people. J Res Health Sci. 2015;15:101–103. [PubMed] [Google Scholar]
- 18.Moaddab SR, Amini K, Haki BK. Determining the drug susceptibility of Mycobacterium tuberculosis strains to the pyrazinamide. Majallah-i pizishki-i Danishgah-i Ulum-i Pizishki va Khadamat-i Bihdashti-i Darmani-i Tabriz. 2015;37:56–62. [Google Scholar]
- 19.Amini S, Hoffner S, Torkaman MR, Hamzehloo G, Nasiri MJ, Salehi M, Kashkooli GS, Shahraki MS, Mohsenpoor M, Soleimanpour S, Mir R. Direct drug susceptibility testing of Mycobacterium tuberculosis using the proportional method: A multicenter study. J Glob Antimicrob Resist. 2019;17:242–244. doi: 10.1016/j.jgar.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 20.Zarei Z, Emami A, Moghadami M, Kashkooli GS, Pirbonyeh N. Molecular characterization of Isoniazid and Rifampicin target genes in multi-drug resistant Mycobacterium tuberculosis isolates from southwest of Iran. Gene Rep. 2017;6:19–25. [Google Scholar]
- 21.Honarvar B, Moghadami M, Emami A, Behbahani AB, Taheri M, Roudgari A, Kashkoli GS, Rezaee M, Farzanfar E, Zaree Z, Goharnejad J. Mycobacterium strain and type of resistance in pulmonary tuberculosis patients: a missed link in Iran’s national tuberculosis plan. Shiraz E-Med J. 2015;16 [Google Scholar]
- 22.Motamedifar M, Ebrahim-Saraie HS, Abadi AR, Moghadam MN. First outcome of MDR-TB among co-infected HIV/TB patients from South-West Iran. Tuberc Respir Dis. 2015;78:253–257. doi: 10.4046/trd.2015.78.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansoori N, Yaseri M, Vaziri F, Douraghi M. Genetic diversity of Mycobacterium tuberculosis complex isolates circulating in an area with high tuberculosis incidence: Using 24-locus MIRU-VNTR method. Tuberculosis. 2018;112:89–97. doi: 10.1016/j.tube.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Zamani S, Haeili M, Nasiri MJ, Imani Fooladi AA, Javadpour S, Feizabadi MM. Genotyping of Mycobacterium tuberculosis isolates from Hormozgan province of Iran based on 15-locus MIRU-VNTR and spoligotyping. Int J Bacteriol. 2016;2016:1–8. doi: 10.1155/2016/7146470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasiri MJ, Rezaei F, Zamani S, Darban-Sarokhalil D, Fooladi AA, Shojaei H, Feizabadi MM. Drug resistance pattern of Mycobacterium tuberculosis isolates from patients of five provinces of Iran. Asian Pac J Trop Med. 2014;7:193–196. doi: 10.1016/S1995-7645(14)60019-5. [DOI] [PubMed] [Google Scholar]
- 26.Karimi S, Mirhendi H, Zaniani FR, Manesh SE, Salehi M, Esfahani BN. Rapid detection of streptomycin-resistant Mycobacterium tuberculosis by rpsL-restriction fragment length polymorphism. Adv Biomed Res. 2017;6:126–132. doi: 10.4103/abr.abr_240_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esfahani BN, Zarkesh FS, Yazdi HR, Radaee T. Detection of embB gene mutations in EMB-resistant Mycobacterium tuberculosis isolates from Isfahan province by PCR-SSCP and direct sequencing. Jundishapur J Microbiol. 2016;9:e39594. [Google Scholar]
- 28.Mohammadi B, Mohajeri P, Rouhi S, Ramazanzadeh R. The relationship between embb306 and embb406 mutations and ethambutol resistant in Mycobacterium tuberculosis isolated from patients in west of Iran. Med J Islam Repub Iran. 2018;32:117. doi: 10.14196/mjiri.32.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohajeri P, Norozi B, Atashi S, Farahani A. Anti tuberculosis drug resistance in west of Iran. J Global Infect Dis. 2014;6:114–117. doi: 10.4103/0974-777X.138506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khosravi AD, Sirous M, Abdi M, Ahmadkhosravi N. Characterization of the most common embCAB gene mutations associated with ethambutol resistance in Mycobacterium tuberculosis isolates from Iran. Infect Drug Resist. 2019;12:579–584. doi: 10.2147/IDR.S196800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khosravi AD, Sirous M, Absalan Z, Tabandeh MR, Savari M. Comparison Of drrA And drrB efflux pump genes expression in drug-susceptible and-resistant Mycobacterium tuberculosis strains isolated from tuberculosis patients In Iran. Infect Drug Resist. 2019;12:3437–44. doi: 10.2147/IDR.S221823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Badie F, Arshadi M, Mohsenpoor M, Gharibvand SS. Drug resistance pattern of Mycobacterium tuberculosis isolates from patients referred to TB reference laboratory in Ahvaz. Osong Public Health Res Perspect. 2016;7:32–35. doi: 10.1016/j.phrp.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khosravi AD, Shahraki AH, Dezfuli SK, Hashemzadeh M, Goodarzi H, Mohajeri P. Genetic diversity of multidrug-resistant Mycobacterium tuberculosis strains isolated from tuberculosis patients in Iran using MIRU-VNTR technique. Kaohsiung J Med Sci. 2017;33:550–557. doi: 10.1016/j.kjms.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khosravi AD, Goodarzi H, Alavi SM, Akhond MR. Application of deletion-targeted multiplex PCR technique for detection of Mycobacterium tuberculosis Beijing strains in samples from tuberculosis patients. Iran J Microbial. 2014;6:330–334. [PMC free article] [PubMed] [Google Scholar]
- 35.Heidary F, Esmaeil Lashgarian H, Karkhane M, Najar Peerayeh S. Molecular detection of isoniazid and rifampin resistance in Mycobacterium tuberculosis isolates from lorestan province, Iran from 2014 to 2017. Arch Clin Infect Dis. 2019;15:e81436. [Google Scholar]
- 36.Farazi A, Sofian M, Zarrinfar N, Katebi F, Hoseini SD, Keshavarz R. Drug resistance pattern and associated risk factors of tuberculosis patients in the central province of Iran. Caspian J Intern Med. 2013;4:785–789. [PMC free article] [PubMed] [Google Scholar]
- 37.Babamahmoodi F, Mahdavi MR, Jalali H, Talebi B, Roshan P, Mahdavi M. Evaluation of gene mutations involved in drug resistance in Mycobacterium tuberculosis strains derived from tuberculosis patients in Mazandaran, Iran, 2013. Int J Mol Cell Med. 2014;3:190–195. [PMC free article] [PubMed] [Google Scholar]
- 38.Sani AT, Shakiba A, Salehi M, Taghanaki HR, Fard SF, Ghazvini K. Epidemiological characterization of drug resistance among Mycobacterium tuberculosis isolated from patients in northeast of Iran during 2012-2013. BioMed Res Int. 2015;2015:1–6. doi: 10.1155/2015/747085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Movahede-Danesh M, Ghazvini K, Heydari A. Evaluation of Antimicrobial Resistance of New Cases of Pulmonary Tuberculosis, in Khorasan, Iran. J Med Bacteriol. 2014;3:45–51. [Google Scholar]
- 40.Hashemi Shahri SM, Fardoust F, Mogharabi Ostad Kalayeh S, Ghenaatpisheh Sanani M. Prevalence of Antibiotic Resistance to Isonicotinylhydrazide and Rifampicin in Culture Positive Pulmonary Tuberculosis Patients from 2014 to 2016 in Zahedan City, Iran. Hosp. Pract Res. 2019;4:57–61. [Google Scholar]
- 41.Shirazinia R, Saadati D, Zeinali E, Mishkar AP. The Incidence and Epidemiology of Tuberculosis in Sistan Region: an Update to Past Researches. Int J Basic Sci Med. 2017;2:189–193. [Google Scholar]
- 42.Vaziri F, Kohl TA, Ghajavand H, Kamakoli MK, Merker M, Hadifar S, Khanipour S, Fateh A, Masoumi M, Siadat SD, Niemann S. Genetic diversity of multi-and extensively drug-resistant Mycobacterium tuberculosis isolates in the capital of Iran, revealed by whole-genome sequencing. J Clin Microbiol. 2019;57:e01477–18. doi: 10.1128/JCM.01477-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Habibnia S, Zaker S, Nasiri MJ, Doustdar F, Ghalavand Z, Ghalami M, Eslami G. Prevalence of multidrug-resistant tuberculosis: a six-year single-center retrospective study in Tehran, Iran. Arch Clin Infect Dis. 2019;14:e82828. [Google Scholar]
- 44.Aghajani J, Saif S, Farnia P, Farnia P, Ghanavi J, Velayati AA. An 8-year study on the prevalence and drug resistance of Mycobacteria in clinical specimens (2011-2018) Clin Epidemiol Glob Health. 2020;8:557–561. [Google Scholar]
- 45.Sakhaee F, Ghazanfari M, Ebrahimzadeh N, Vaziri F, Jamnani FR, Davari M, Gharibzadeh S, Mandjin FH, Fateh A, Siadat SD. A comparative study of phenotypic and genotypic first-and second-line drug resistance testing of Mycobacterium tuberculosis. Biologicals. 2017;49:33–38. doi: 10.1016/j.biologicals.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Khanipour S, Ebrahimzadeh N, Masoumi M, Sakhaei F, Alinezhad F, Safarpour E, Fateh A, Nematollahi AN, Tasbiti AH, Zolfaghari MR, Bahrmand AR. Haarlem 3 is the predominant genotype family in multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis in the capital of Iran: a 5-year survey. J Glob Antimicrob Resist. 2016;5:7–10. doi: 10.1016/j.jgar.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 47.Imani Fooladi AA, Babak F, Fazlollah MS, Nematollah JJ. Rapid detection of MDR–Mycobacterium tuberculosis using modified PCR-SSCP from clinical Specimens. Asian Pac J Trop Biomed. 2014;4:S165–170. doi: 10.12980/APJTB.4.2014C1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tasbiti AH, Yari S, Ghanei M, Shokrgozar MA, Fateh A, Bahrmand A. Low Levels of Extensively Drug-resistant Tuberculosis among Multidrug Resistant Tuberculosis Isolates and Their Relationship to Risk Factors: Surveillance in Tehran, Iran; 2006 to 2014. Osong Public Health Res Perspect. 2017;8:116–123. doi: 10.24171/j.phrp.2017.8.2.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharifipour E, Nasiri M, Farnia P, Mozafari M, Irani S. Evaluation of molecular diversity of Mycobacterium tuberculosis strains by polymorphisms in RD Regions. J Mycobac Dis. 2014;4:2161–1068. [Google Scholar]
- 50.Bahrami S, Bahrmand AR, Safarpour E, Masoumi M, Saifi M. Detection of ethambutol-resistant associated mutations in Mycobacterium tuberculosis isolates from Iran using multiplex allele-specific PCR. J Med Microbiol Infec Dis . 2013;1:41–45. [Google Scholar]
- 51.Pooideh M, Jabbarzadeh I, Ranjbar R, Saifi M. Molecular epidemiology of Mycobacterium tuberculosis isolates in 100 patients with tuberculosis using pulsed field gel electrophoresis. Jundishapur J Microbiol. 2015;8:e18274. doi: 10.5812/jjm.8(5)2015.18274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varahram M, Nasiri MJ, Farnia P, Mozafari M, Velayati AA. A retrospective analysis of isoniazid-monoresistant tuberculosis: among Iranian pulmonary tuberculosis patients. Open Microbiol J. 2014;8:1–5. doi: 10.2174/1874285801408010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bialvaei AZ, Asgharzadeh M, Aghazadeh M, Nourazarian M, Kafil HS. Challenges of tuberculosis in Iran. Jundishapur J Microbiol. 2017;10:e37866. [Google Scholar]
- 54.LoBue PA, Moser KS. Use of isoniazid for latent tuberculosis infection in a public health clinic. Am J Respir Crit Care Med. 2003;168:443–7. doi: 10.1164/rccm.200303-390OC. [DOI] [PubMed] [Google Scholar]
- 55.Feyisa SG, Abdurahman AA, Jimma W, Chaka EE, Kardan-Yamchi J, Kazemian H. Resistance of Mycobacterium tuberculosis strains to Rifampicin: A systematic review and meta-analysis. Heliyon. 2019;5:e01081. doi: 10.1016/j.heliyon.2018.e01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miotto P, Zhang Y, Cirillo DM, Yam WC. Drug resistance mechanisms and drug susceptibility testing for tuberculosis. Respirology. 2018;23:1098–113. doi: 10.1111/resp.13393. [DOI] [PubMed] [Google Scholar]