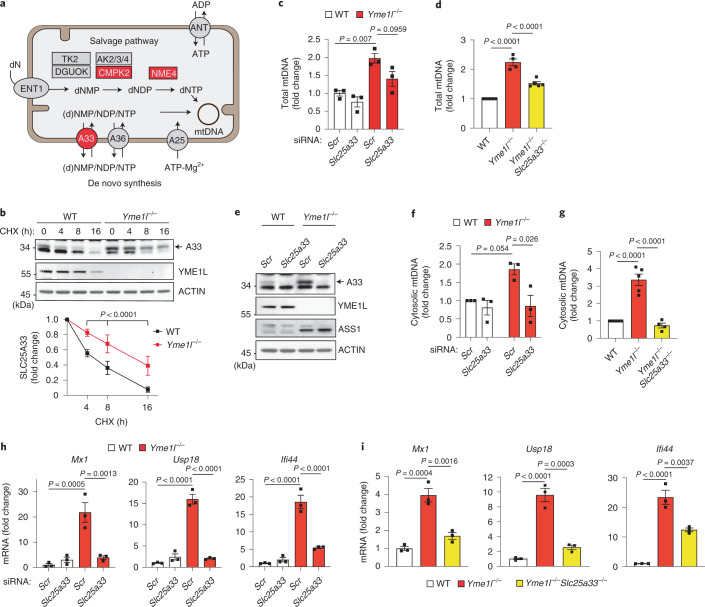
Fig. 3. Proteolysis of SLC25A33 by YME1L controls mtDNA-dependent innate immunity.
a, Nucleotide synthesis via mitochondrial salvage or cytosolic de novo pathway. Components that affect the metabolism of mtDNA and that accumulate in Yme1l−/− cells are highlighted in red. b, Cycloheximide (CHX) treatment of MEFs for the indicated time. Quantification of SLC25A33 levels is shown in the lower panel (mean ± SD; n = 7 independent cultures). c,d, Total mtDNA level monitored by qPCR (Cytb) in WT and Yme1l−/− MEFs treated with the indicated siRNAs (Slc25a33 no. 1) (n = 3 independent cultures) (c) or in WT, Yme1l−/− and Yme1l−/−Slc25a33−/− MEFs (n = 5 independent cultures for WT and Yme1l−/−Slc25a33−/−; n = 4 independent cultures for Yme1l−/− (d). e, Immunoblot analysis of WT and Yme1l−/− MEFs treated with the indicated siRNAs (Slc25a33 no. 1) (representative blot from n = 3 independent cultures). f,g, mtDNA levels in cytosolic fractions monitored by qPCR (Cytb) in WT and Yme1l−/− MEFs treated with the indicated siRNAs (Slc25a33 no. 1) (n = 3 independent cultures) (f) or in WT, Yme1l−/− and Yme1l−/−Slc25a33−/− MEFs (n = 5 independent cultures for WT and Yme1l−/−; n = 4 independent cultures for Yme1l−/−Slc25a33−/−) (g). h,i, ISG expression in WT and Yme1l−/− MEFs treated with the indicated siRNAs (Slc25a33 no. 2) (n = 3 independent cultures) (h) or in WT, Yme1l−/− and Yme1l−/−Slc25a33−/− MEFs (n = 3 independent cultures) (i). P values calculated using two-tailed multiple t-test with Holm–Sidak method to correct for multiple comparisons (b), two-way ANOVA with Tukey’s multiple comparison test (c,f,h) or one-way ANOVA with Tukey’s multiple comparison test (d,g,i). Data (except b) are means ± s.e.m.