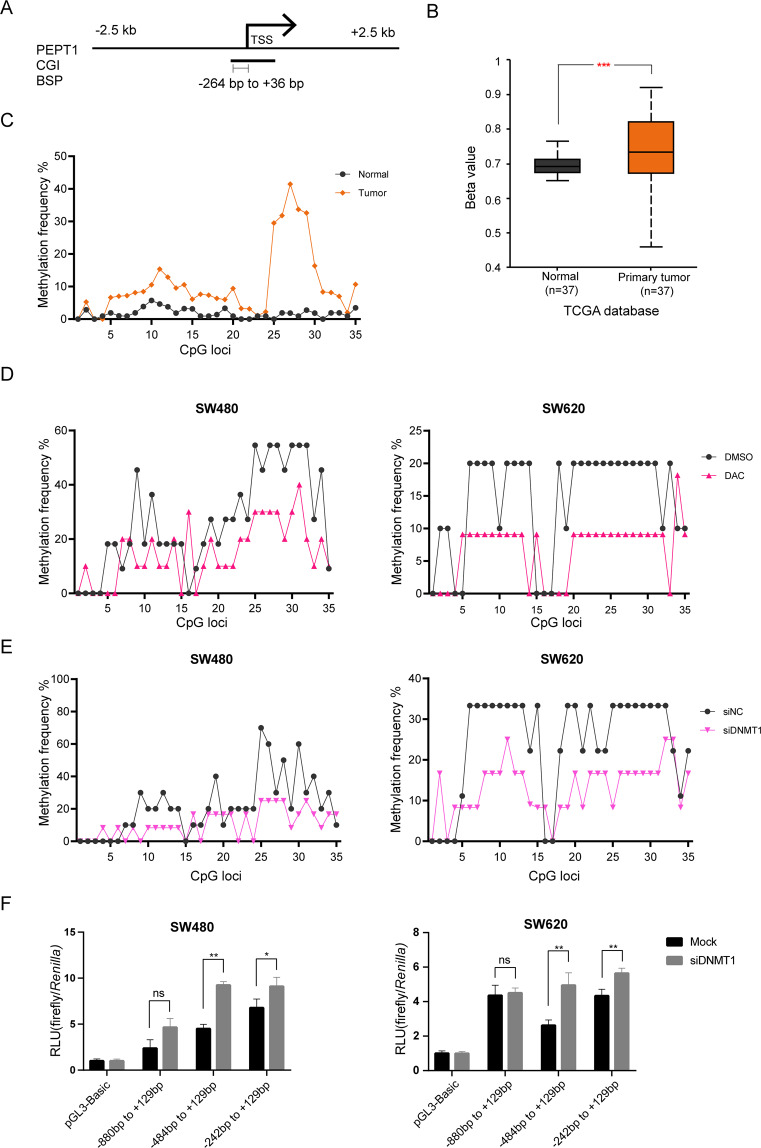
Fig. 3. Repression of PEPT1 expression via DNMT1 mediated DNA hypermethylation.
A PEPT1 promoter region (±2.5 kb) around 5’ regions adjacent to the TSS. CGI, CpG islands. BSP, bisulfite-sequencing PCR. B DNA methylation data of PEPT1 promoter in cancerous and adjacent non-tumorous tissues from the TCGA database (***P < 0.001). C BSP analysis of PEPT1 CGI in cancerous and adjacent non-tumorous tissues (n = 11). Methylation percentages of the 35 CpG loci in the sequenced region were calculated. 11 pairs of CRC tissues belong to tissues in Fig. 1B (CRC17, 19, 22, 26, 41, 47, 49, 50, 51, 52, 54). The y axis indicates the average methylation percentage of each CpG site calculated from patient tissues with PEPT1 repression. D–E Calculation of methylation percentages of PEPT1 promoter after DAC and siDNMT1 treatment in CRC cells. F Luciferase assay in DNMT1 knockdown CRC cells. Mock, cells transfected with negative control siRNA and PEPT1 promoter constructs. Si-DNMT1, cells transfected with siDNMT1#2 and PEPT1 promoter constructs. Data are shown as means ± SD, two-tailed unpaired t-test, *P < 0.05, **P < 0.01.