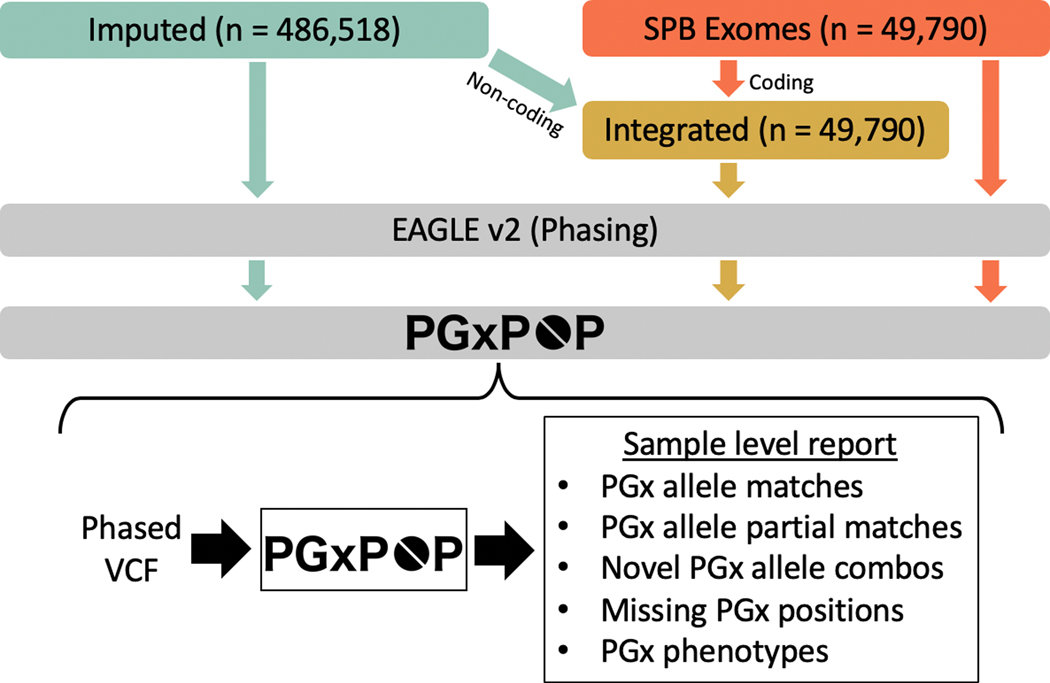
Figure 1.
Analysis workflow. Our analysis comprises three data types, data imputed from genotype arrays, exome sequencing data, and an integrated call set that combines both. We phase all datasets using statistical phasing with Eagle2. We then generate pharmacogenetic alleles for all samples using PGxPOP and generate a report of the matching star allele, the variants contributing to that call, and the resulting phenotype.