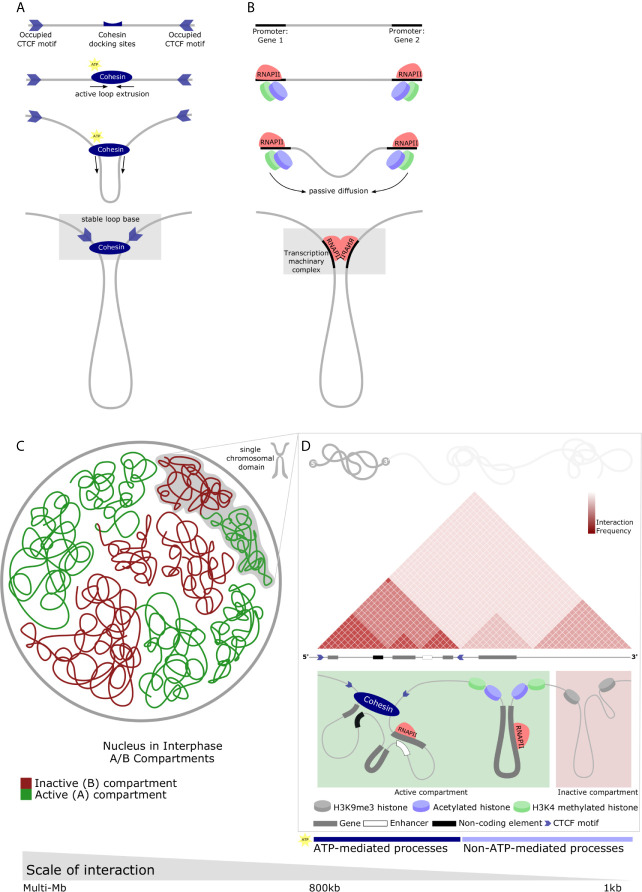
Figure 1.
Diverse chromatin structures across the range of nuclear scale and the main proposed regulatory mechanisms which underlie chromatin structure. (A) CTCF-mediated looping forms TADs. Directional CTCF motifs across the genome are occupied by CTCF in relation to loading/unloading dynamics of nuclear proteins. Similarly, Cohesin binds chromatin (top panel) and extrudes chromatin through its closed ring structure to form a loop (middle panels). Extrusion continues bidirectionally until convergent CTCF occupied sites are encountered. Heterodimerization of CTCFs, Cohesin, and various other proteins form a stable loop anchor complex (bottom panel) and are the basis of most stable TAD structures. (B) RNAPII-mediated looping forms small chromatin loops. TFs bind motifs in the regulatory elements of genes along with RNAPII (top-middle panel). Passive diffusion and random motion which are not ATP-mediated bring these protein structures into proximity with one-another (middle-bottom panel) and, by means of affinity-interactions, chromatin loops form between regulatory elements (bottom panel). These protein-protein interactions may result in liquid-liquid phase separation and the relatively dynamic recruitment of transcriptional machinery, creating a permissive environment for rapid and robust transcription. (C) Nuclear compartments are complex chromatin structures. In interphase, the nucleus is divided into A and B compartments. These domains span multiple megabases and comprise active (gene-dense, euchromatin) and inactive (gene-scarce, heterochromatin) possibly mediated by liquid-liquid phase separation. Green - active or A compartment; red - inactive or B compartment. (D) Multiple different chromatin loop structures are contained within single chromosomes which contribute to A or B compartments. Individual chromosomes can contain contiguous regions of active or inactive chromatin which represent the interactions of various elements of the genome facilitating functional regulation. These chromatin interactions are depicted by a schematic of a Hi-C map demonstrating a small area of a single chromosome in interphase. Triangular regions of varying red intensity represent the likelihood of interactions between chromatin regions in a population of cells. TAD with boundaries occupied by convergent CTCF motifs insulate areas of the genome (left). Similarly, the active compartment contribution of this representative chromosome may house RNAPII-mediate loops mediating interactions between genes, or genes and non-coding regulatory elements (as described in B) (middle). The inactive compartment contribution may be mediated by protein-mediated liquid-liquid phase separation to form repressive compartmental domains in gene-scarce heterochromatin (right). These processes can be either ATP- or non-ATP-mediated.