Figure 2.
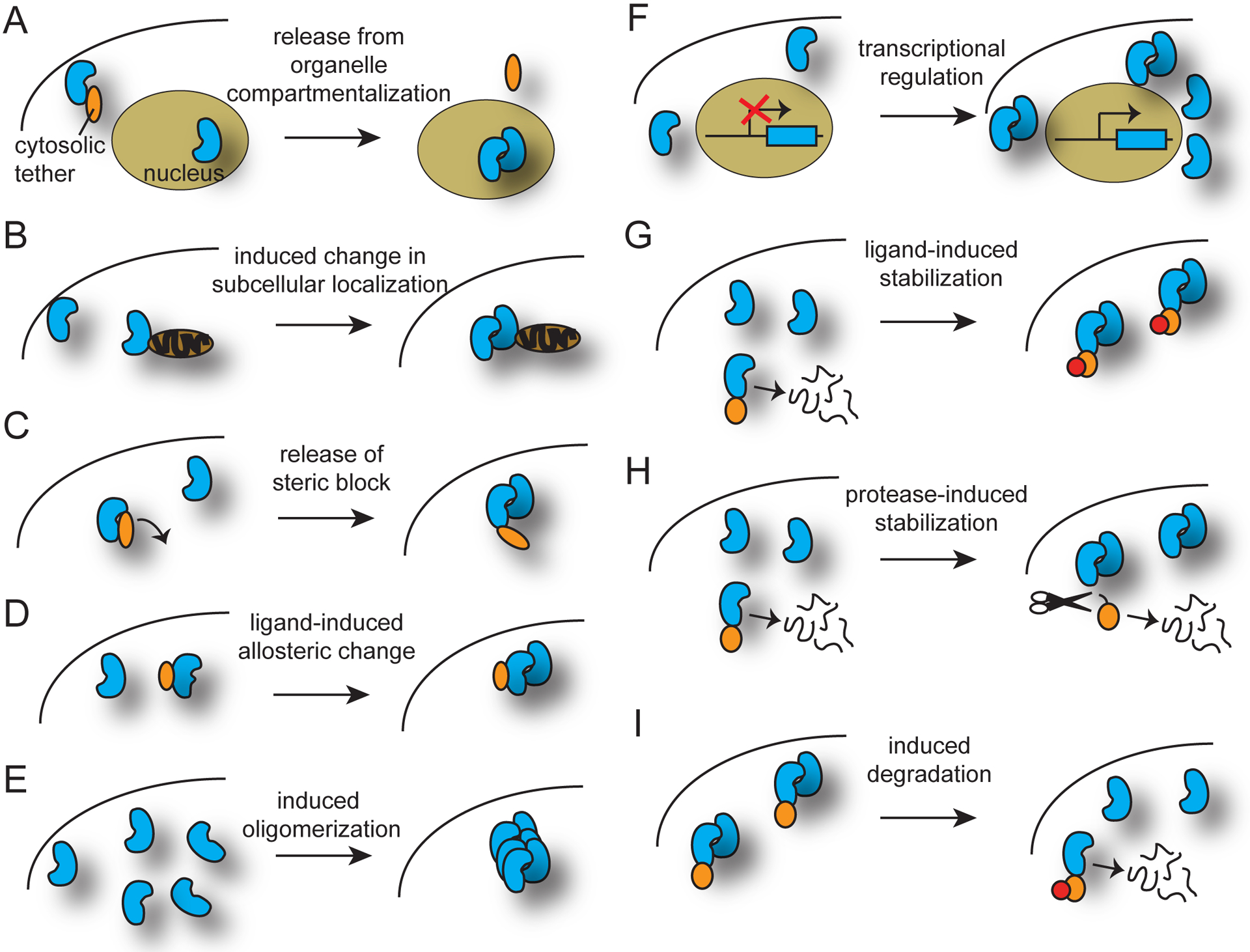
Additional regulation layered upon inducible dimerization systems. Inducible dual system approaches are divided into two categories, regulation of system component properties (protein localization, activity, or binding) without altering actual protein levels and (A-E) and regulation of actual protein abundance (F-I). (A) Release from organelle compartmentalization. In the example shown, a dimerizer component is prevented from entering its functional organelle (the nucleus) through anchoring to a cytosolic tether. The protein can be inducibly released through several mechanisms, including use of light or ligand-activated dimerization systems and inducible protease cleavage. (B) Induced change in subcellular localization. This concept is similar to (A), but does not involve compartmentalization within an organelle. (C) Release of steric block. In the example shown, a light- or ligand-sensitive protein acts to prevent association of a dimerizer-dependent split protein in the uninduced state. With the inducer, the steric block is removed and the protein can associate. (D) Ligand-induced allosteric change. (E) Induced oligomerization. (F) Inducible transcriptional regulation. A light- or ligand-induced transcriptional system is layered onto a dimerization system. (G) Ligand-induced protein stabilization. A destabilized ligand-binding domain is added to one or more components, resulting in protein degradation in the absence of inducer. (H) Protease-induced protein stabilization. Similar to (G), a degron is attached to a component, resulting in constitutive degradation unless a protease is active, removing the degron. The cleavage can be regulated by a dimerized split protease, a photosensitive cleavage site (e.g., PhoCl or AsLOV-caged cleavage site), or a protease inhibitor. (I) Inducible protein degradation. A protein is fused to a domain that can be conditionally targeted to the proteasome (with addition of an inducer).
