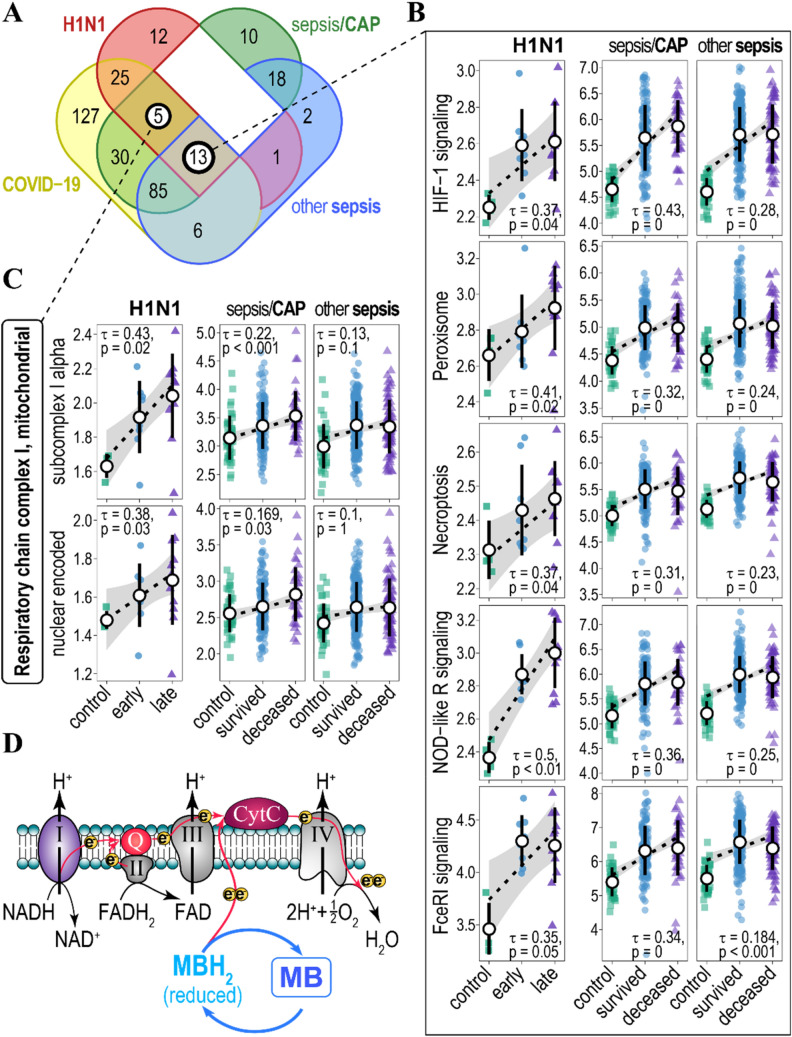
Figure 2.
Association of gene ensemble noise with H1N1 infection phase and sepsis mortality. (A) Venn diagram of KEGG- and CORUM-annotated biological pathways/protein complexes for which gene ensemble noise associates positively (increases) and significantly with disease state (mild/severe for the COVID-19, healthy/early/late for the H1N1, and healthy/survived/deceased for sepsis/CAP and other sepsis patients). (B) Plots of gene ensemble noise for genes involved in HIF-1 signalling, peroxisome, necroptosis, NOD-like receptor, and Fc epsilon RI signalling pathways. Pathways were annotated by KEGG. Kendall tau, and FDR- (H1N1 patients) and Bonferroni- (sepsis patients) adjusted p-values are indicated. Rank-based regression trend lines and 95% confidence bands of gene ensemble noise association with the state of disease are shown. Black circles and whiskers indicate means and standard deviations. (C) Plots of gene ensemble noise for genes encoding CORUM-annotated subunits of mitochondrial respiratory chain complex I (subcomplex I alpha—top panel and nuclear-encoded subunits—bottom panel). Rank-based regression trend lines and 95% confidence bands of gene ensemble noise association with the state of the disease are shown. (D) Methylene Blue (MB) acts as an alternative electron donor to the electron transport chain (red arrows) by shuttling between redox states (MB—MBH2) and, thus, bypassing respiratory chain complex I. Respiratory chain complex I-IV and their substrates are indicated, Q—coenzyme Q10, CytC—cytochrome C. Electrons are indicated as yellow circles.