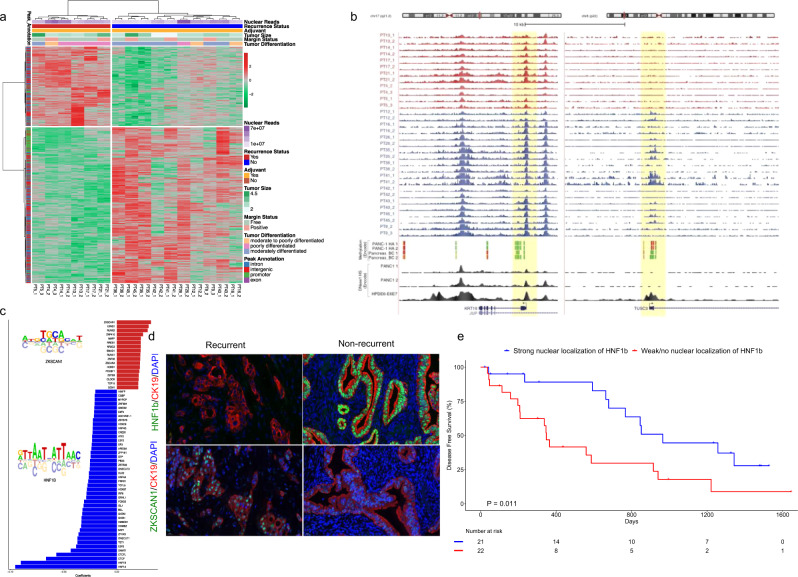
Fig. 1. Differential chromatin accessibility signature and transcription factor nuclear localization predict prognosis of resected PDAC.
a Differential ATAC-seq peaks in recurrent versus non-recurrent patients (Source data are provided as a Source Data file). b Upper panel depicts genome browser tracks showing ATAC-seq peaks at KRT19 and TUSC3 gene loci for duplicate samples from individual patients (PT); lower panel shows corresponding ENCODE methylation signals from Panc-1 and a normal pancreatic (BC) cell lines, (green denotes hypomethylation and red denotes hypermethylation) and DHS peak from Panc-1 and HPDE6-E6E7 cell lines. Highlighted yellow regions denote the promoter peaks of both the gene loci. All the track lines have the same Y axis limits and that the peak height is normalized by the total read depth of each sample. c Regression coefficients showing enrichment of transcription factor-binding sites in recurrent (red) and non-recurrent (blue) patients. d Representative images of TMA staining of HNF1b (upper panel) and ZKSCAN1 (lower panel) in recurrent and non-recurrent patients (TMA n = 43 with three replicated cores in each patient), Scale bars are 20 μM as displayed at the left bottom corners of all the micrographs. e Kaplan–Meier curve of the patients with and without nuclear localization of HNF1b (log-rank (Mantel–Cox) test P = 0.011, HR 2.64, 95% CI 1.213 to 5.775). (Source data are provided as a Source Data file).