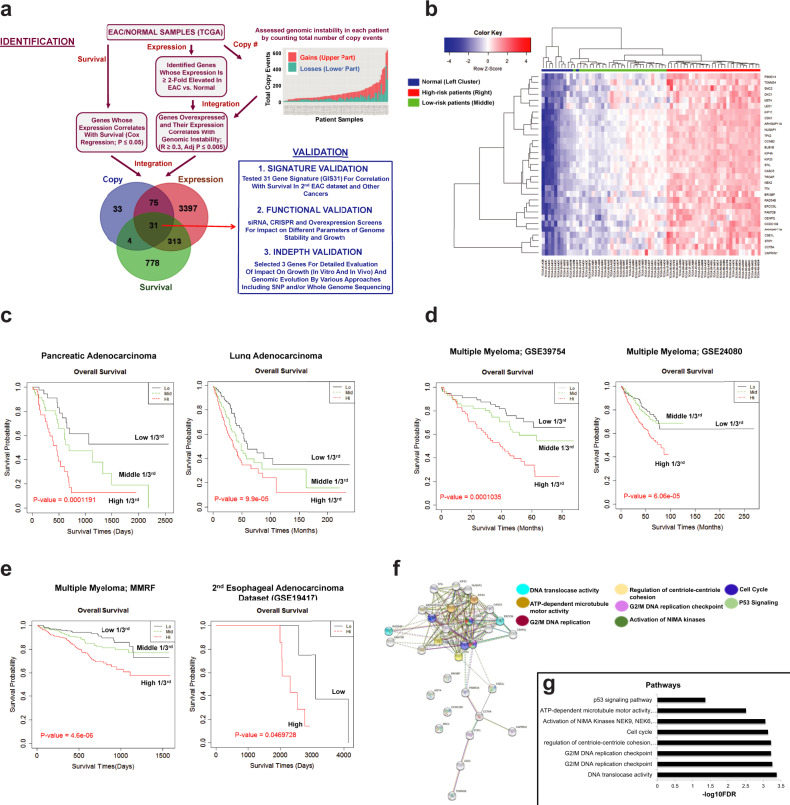
Fig. 1. Identification of drivers of genomic evolution in EAC.
a Experimental design. Stepwise process involved in the identification and validation of genes. Identification: (1) assessed gene expression in 11 normal and 88 EAC patient samples in TCGA dataset and identified genes that were overexpressed in EAC; (2) asessed genomic instability in patient samples by counting total number of copy events in each patient. Total copy number events in each patient are shown as bar graph in Fig. 1a; each bar shows amplifications (red or upper part of bar) and deletions (green or lower part of the bar). Genomic instability data were then integrated with expression data to identify genes that were overexpressed in EAC and whose expression correlated with genomic instability; (3) evaluated resulting genes for correlation with survival. This led to identification of 31 genes that were overexpressed in EAC and whose expression correlated with genomic instability and overall survival in EAC patients in TCGA dataset. Validation: describes plan for functional validation of genes: (1) the expression of 31 gene signature was tested for correlation with survival in a second EAC dataset and in other cancers; (2) conducted siRNA, CRISPR/Cas9, and overexpression screens for impact on different parameters of genome stability and growth; (3) selected three genes for detailed evaluation of impact on genomic evolution by various approaches including SNP and/or whole genome sequencing; (4) evaluation in SCID mice. b Heat map shows that expression of 31 genes is high in EAC patients with poor overall survival (red bar) and low in patients with relatively better survival (green bar) (in TCGA dataset), and also low in normal samples (blue bar). c–e Elevated expression of 31 gene signatures (identified in TCGA patient dataset) correlated with poor overall survival in pancreatic and lung adenocarcinoma (c), multiple myeloma datasets GSE39754 and GSE24080 (d), and multiple myeloma MMRF (Multiple Myeloma Research Foundation) dataset and second esophageal adenocarcinoma dataset (GSE19417) (e). f Interconnectivity among the GIS31 genes (based on STRING networks) is shown by lines and the shared functional roles of these genes in different pathways are indicated by different colors. g The pathways shown in f are also shown as bar graphs.