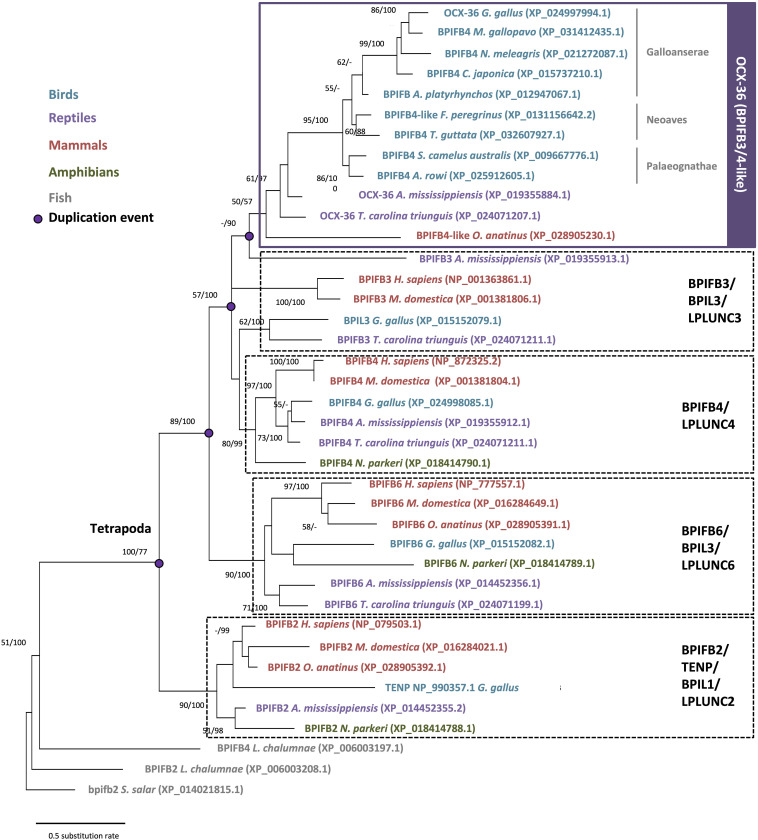
FIGURE 6.
Phylogenetic reconstruction of OCX-36 and its orthologs and paralogs in vertebrates. For phylogeny multiple alignment was performed using ClustalW and Gblocks (www.phylogeny.fr). Model of protein evolution used is JTT + G (MEGAX v10.1.8; https://www.megasoftware.net/). Topology of the tree corresponds to maximum likelihood method with 100 repetitions (MEGAX v10.1.8). Bootstrap values from maximum likelihood/Bayesian inference are indicated at each node when value is >50.