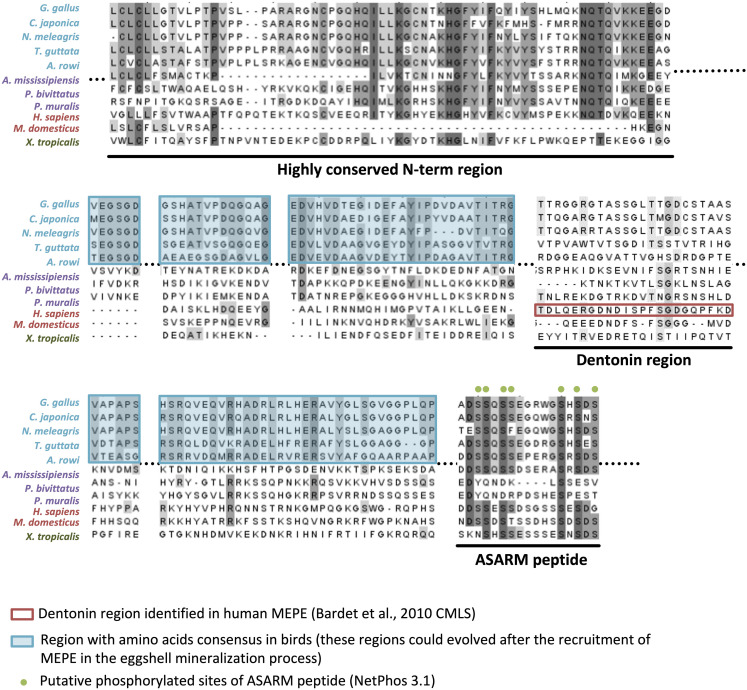
FIGURE 7.
Multiple alignment of OC-116/MEPE proteins in tetrapods. The alignment was built with ClustalW (www.phylogeny.fr) and edited with Jalview v2.11.1.4 (https://www.jalview.org/). Gray colors of letters correspond to percentage of identity (dark gray > 80% identity, medium gray > 60% identity, and light gray > 40% identity). The dentonin region identified in human MEPE, and regions with amino acid consensus in birds, are, respectively, framed in red and blue. Putative phosphorylated sites of the ASARM (acidic, serine, and aspartic acid-rich motif) peptide are indicated by green dots above the alignment. Sequences aligned are from Gallus gallus (AAF00982.3), Coturnix japonica (XP_015716951.1), Numida meleagris (XP_021251779.1), Taeniopygia guttata (XP_030127798.1), Apteryx rowi (XP_025920573.1), Alligator mississippiensis (XP_019343676.1), Python bivittatus (XP_025029286.1), Podarcis muralis (XP_028598766.1), Homo sapiens (XP_006714341.1), Monodelphis domestica (XP_007495981.1), and Xenopus tropicalis (XP_002938672.2).