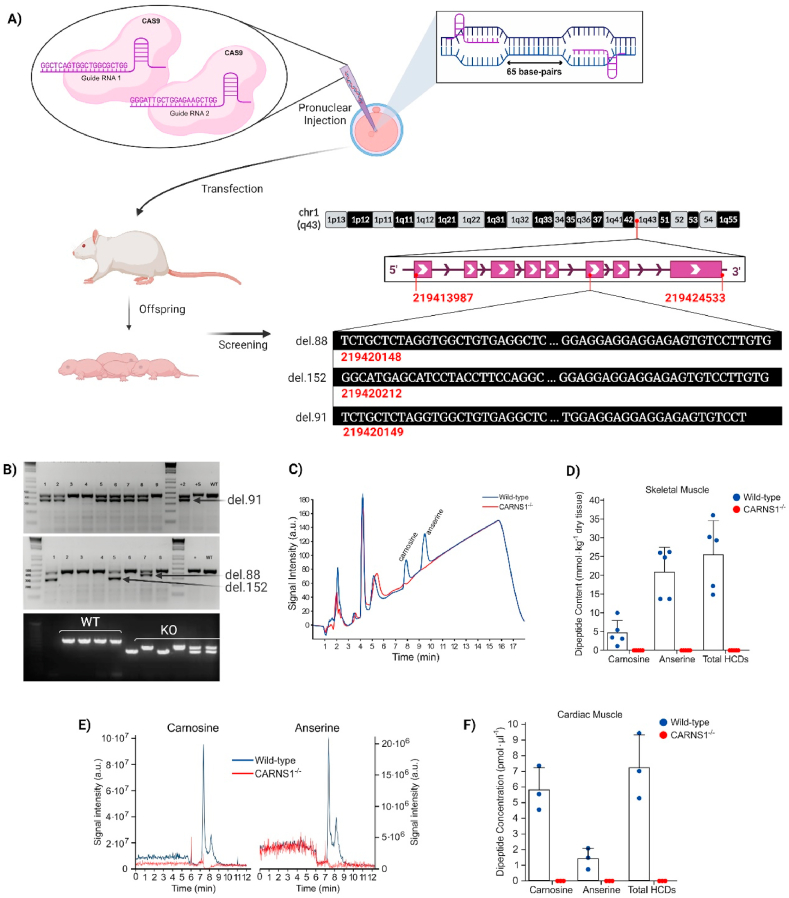
Fig. 1.
A) Two CRISPR guides were designed targeting exon 6 of the CARNS1 gene aiming at large deletions and frameshift. They were injected along with Cas9 into pronuclei of rat embryos. B) Offspring were screened via gene sequencing and 3 mutated alleles were identified. The red numbers display their chromosome positions, the DNA sequences in white display their deleted sequences, along with the size of the deletions. PCR genotyping confirmed amplicon sizes (upper gel images were carried out at Transposagen Inc.). In-house genotyping of rats bred to homozygosity confirmed transmission of mutated alleles. C and D) Representative chromatograms and HCD quantification via HPLC in skeletal muscle samples of WT and CARNS1−/− rats. Signals below the limits of detection were deemed zero. E and F) Representative SRM chromatograms and histidine-containing dipeptide quantification via HPLC-ESI + -MS/MS in cardiac muscle samples in WT and CARNS1−/− rats. The signal to noise ratio of ≥3 was used as the detection criteria for the adducts and a S/N ≥ 7 was used as the quantification criteria. Values below this threshold were deemed zero. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)