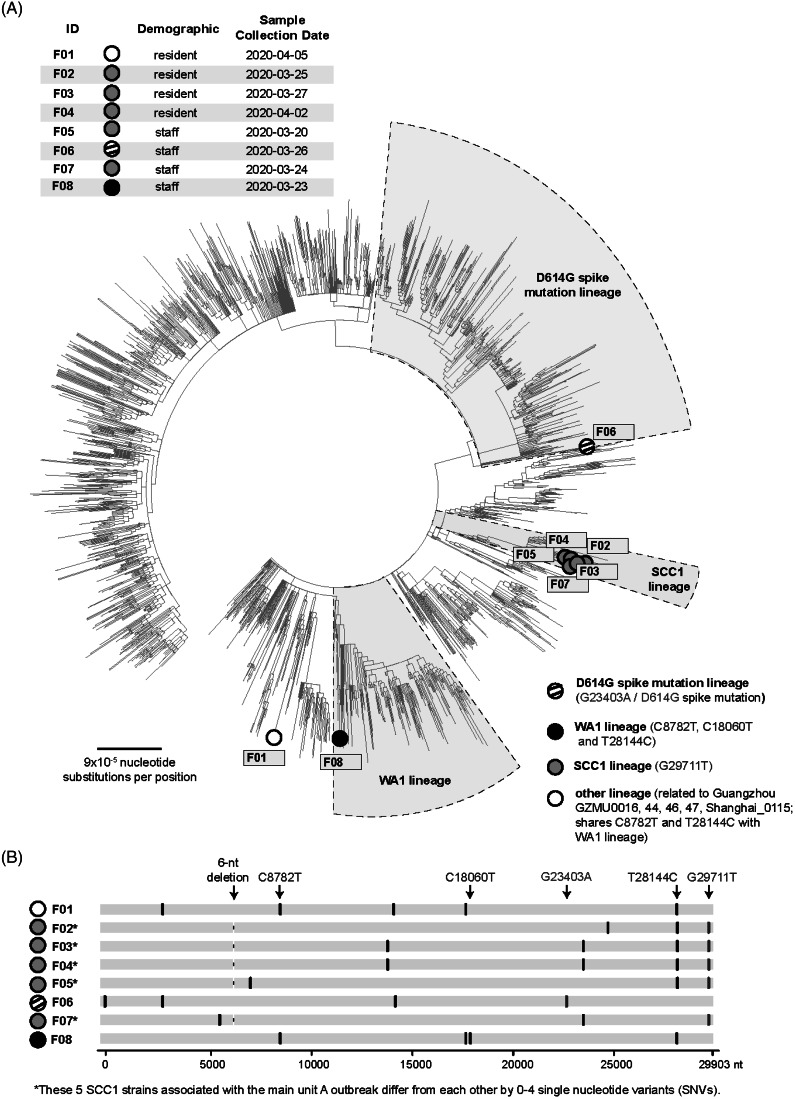
Fig. 2.
Viral lineages in the facility outbreak (n = 8) from March 22 to May 4, 2020. (A) Phylogenetic tree of the 8 facility genomes along with all SARS-CoV-2 US genomes deposited in the GISAID reference database as of May 28, 2020 and all non-US global genomes deposited as of Mar 23, 2020. The 5 strains associated with the main unit A outbreak, 3 from patients and 2 from providers, are all found to map to the SCC1 lineage. (B) Multiple sequence alignment of the 8 facility viruses. The viral genomes are aligned to the reference SARS-CoV-2 Wuhan-1 strain. Key signature SNVs relative to the reference strain are marked by lineage; other SNVs are shown in black. A 6-nt deletion is present in each of the 5 F outbreak strains. Note. F, Facility; SNVs, single nucleotide variants; WA, Washington; SCC, Santa Clara County; GISAID, Global Initiative for Sequencing of All Influenza Data (expanded to include SARS-CoV-2; nt, nucleotide).