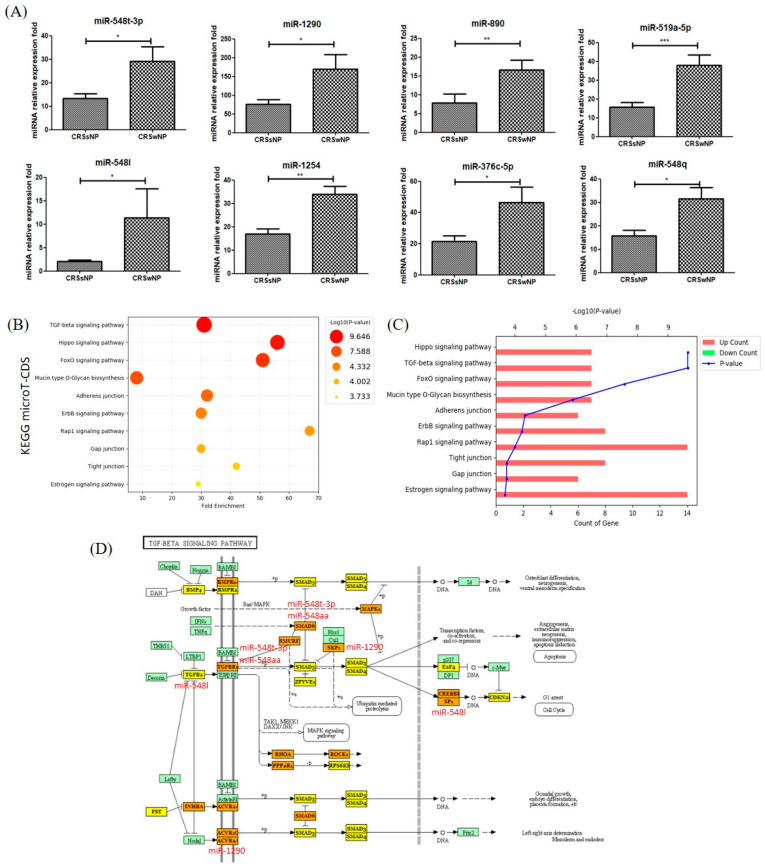
Figure 4.
Top 10 KEGG biological pathways revealed by analyzing the target miRNAs between CRSsNP and CRSwNP NLF-EVs using DIANA tools. (A) Overexpressed miRNAs in NLF-EVs of CRS patients compared to HCs after adjustment of outlier values according to the Chauvenet criterion (Student’s t-test; * p < 0.05; ** p < 0.01; *** p < 0.001). (B) KEGG enrichment analysis of all DEGs with |FC| > 2 (cutoff). (C) Pathway enriched by up- and downregulated genes and sorted by the value of −log10(p-value). (D) Diagram of the TGF-β signaling pathway. Six dysregulated miRNAs (miR-1290, miR-548q, miR-548l, miR-1254, miR-890, and miR-548aa/548t) were associated with the TGF-β signaling pathway by possibly regulating their potential gene targets. KEGG, Kyoto Encyclopedia Gene and Genome; miRNA, microRNA; CRSsNP, chronic rhinosinusitis without nasal polyps; CRSwNP, chronic rhinosinusitis with nasal polyps; NLF, nasal lavage fluid; EV, extracellular vesicle; CRS, chronic rhinosinusitis; HC, healthy control; DEG, differentially expressed gene; FC, fold change; TGF-β, transforming growth factor-beta.